Utilizing Stable Gene-Edited Knockout Pools for Genetic Screening and Engineering in Chinese Hamster Ovary Cells
- PMID: 40376717
- PMCID: PMC12082383
- DOI: 10.1002/biot.70033
Utilizing Stable Gene-Edited Knockout Pools for Genetic Screening and Engineering in Chinese Hamster Ovary Cells
Abstract
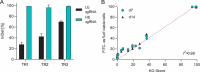
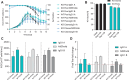
Chinese hamster ovary (CHO) cells are the primary host for biopharmaceutical production. To meet increasing demands for productivity, quality, and complex molecule expression, genetic engineering, particularly clustered regularly interspaced short palindromic repeats (CRISPR)-mediated gene knockout (KO), is widely used to optimize host cell performance. However, systematic screening of KO targets remains challenging due to the labor-intensive process of generating and evaluating individual clones. In this study, we present a robust, high-throughput CRISPR workflow using stable KO pools in CHO cells. These pools maintain genetic stability for over 6 weeks, including in multiplexed configurations targeting up to seven genes simultaneously. Compared to clonal approaches, KO pools reduce variability caused by clonal heterogeneity and better reflect the host cell population phenotype. We demonstrate the utility of this approach by reproducing the beneficial phenotypic effects of fibronectin 1 (FN1) KO, specifically prolonged culture duration and improved late-stage viability in fed-batch processes. This workflow enables efficient identification and evaluation of promising KO targets without the need to generate and test large numbers of clones. Overall, screening throughput is increased 2.5-fold and timelines are compressed from 9 to 5 weeks. This provides a scalable, efficient alternative to traditional clonal screening, accelerating discovery for CHO cell line engineering for biopharmaceutical development.
Keywords: CHO; clonal heterogeneity; genetic engineering; host cell line engineering; stable CHO pools.
© 2025 The Author(s). Biotechnology Journal published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
CRISPR/Cas12a-mediated CHO genome engineering can be effectively integrated at multiple stages of the cell line generation process for bioproduction.Biotechnol J. 2021 Apr;16(4):e2000308. doi: 10.1002/biot.202000308. Epub 2021 Jan 25. Biotechnol J. 2021. PMID: 33369118
-
High throughput, efficacious gene editing & genome surveillance in Chinese hamster ovary cells.PLoS One. 2019 Dec 19;14(12):e0218653. doi: 10.1371/journal.pone.0218653. eCollection 2019. PLoS One. 2019. PMID: 31856197 Free PMC article.
-
CRISPR/Cas9-Mediated Knockout of MicroRNA-744 Improves Antibody Titer of CHO Production Cell Lines.Biotechnol J. 2019 May;14(5):e1800477. doi: 10.1002/biot.201800477. Epub 2019 Apr 15. Biotechnol J. 2019. PMID: 30802343
-
CRISPR Technologies in Chinese Hamster Ovary Cell Line Engineering.Int J Mol Sci. 2023 May 2;24(9):8144. doi: 10.3390/ijms24098144. Int J Mol Sci. 2023. PMID: 37175850 Free PMC article. Review.
-
CRISPR/Cas9-mediated genome engineering of CHO cell factories: Application and perspectives.Biotechnol J. 2015 Jul;10(7):979-94. doi: 10.1002/biot.201500082. Epub 2015 Jun 9. Biotechnol J. 2015. PMID: 26058577 Review.
References
-
- Ha T. K., Hansen A. H., Kildegaard H. F., and Lee G. M., “Knockout of Sialidase and Pro‐Apoptotic Genes in Chinese Hamster Ovary Cells Enables the Production of Recombinant human Erythropoietin in Fed‐Batch Cultures,” Metabolic Engineering 57 (2020): 182–192, 10.1016/j.ymben.2019.11.008. - DOI - PubMed
-
- Lin P.‐C., Liu R., Alvin K., et al., “Improving Antibody Production in Stably Transfected CHO Cells by CRISPR‐Cas9‐Mediated Inactivation of Genes Identified in a Large‐Scale Screen With Chinese Hamster‐Specific siRNAs,” Biotechnology Journal 16 (2021): 2000267, 10.1002/biot.202000267. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

