Genome-Wide Analysis of DtxR and HrrA Regulons Reveals Novel Targets and a High Level of Interconnectivity Between Iron and Heme Regulatory Networks in Corynebacterium glutamicum
- PMID: 40376914
- PMCID: PMC12327846
- DOI: 10.1111/mmi.15376
Genome-Wide Analysis of DtxR and HrrA Regulons Reveals Novel Targets and a High Level of Interconnectivity Between Iron and Heme Regulatory Networks in Corynebacterium glutamicum
Abstract
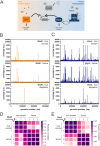
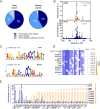
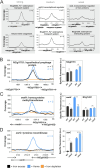
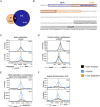
Iron is vital for most organisms, serving as a cofactor in enzymes, regulatory proteins, and respiratory cytochromes. In Corynebacterium glutamicum , iron and heme homeostasis are tightly interconnected and controlled by the global regulators DtxR and HrrA. While DtxR senses intracellular Fe2+, HrrSA is activated by heme. This study provides the first genome-wide analysis of DtxR and HrrA binding dynamics under varying iron and heme conditions using chromatin affinity purification and sequencing (ChAP-Seq). We revealed 25 novel DtxR targets and 210 previously unrecognized HrrA targets. Among these, metH, encoding homocysteine methyltransferase, and xerC, encoding a tyrosine recombinase, were bound by DtxR exclusively under heme conditions, underscoring condition-dependent variation. Activation of metH by DtxR links iron metabolism to methionine synthesis, potentially relevant for the mitigation of oxidative stress. Beyond novel targets, 16 shared targets between DtxR and HrrA, some with overlapping operator sequences, highlight their interconnected regulons. Strikingly, we demonstrate the significance of weak ChAP-Seq peaks that are often disregarded in global approaches, but feature an impact of the regulator on differential gene expression. These findings emphasize the importance of genome-wide profiling under different conditions to uncover novel targets and shed light on the complexity and dynamic nature of bacterial regulatory networks.
Keywords: chromatin affinity purification and sequencing; heme; homeostasis; iron; transcription factors.
© 2025 The Author(s). Molecular Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Control of heme homeostasis in Corynebacterium glutamicum by the two-component system HrrSA.J Bacteriol. 2011 Mar;193(5):1212-21. doi: 10.1128/JB.01130-10. Epub 2011 Jan 7. J Bacteriol. 2011. PMID: 21217007 Free PMC article.
-
The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum.BMC Genomics. 2006 Feb 9;7:21. doi: 10.1186/1471-2164-7-21. BMC Genomics. 2006. PMID: 16469103 Free PMC article.
-
Regulatory networks of FUR and NtcA are intertwined by transcriptional regulators, two-component systems, serine/threonine kinases, and sigma factors in Anabaena sp. PCC 7120.mSystems. 2025 Jul 22;10(7):e0037325. doi: 10.1128/msystems.00373-25. Epub 2025 Jun 25. mSystems. 2025. PMID: 40558093 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
References
-
- Ando, M. , Manabe Y. C., Converse P. J., et al. 2003. “Characterization of the Role of the Divalent Metal Ion‐Dependent Transcriptional Repressor MntR in the Virulence of <styled-content style="fixed-case"> Staphylococcus aureus </styled-content> .” Infection and Immunity 71, no. 5: 2584–2590. 10.1128/iai.71.5.2584-2590.2003. - DOI - PMC - PubMed
-
- Bailey, T. L. , and Elkan C.. 1994. “Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers.” Proceedings of the International Conference on Intelligent Systems for Molecular Biology 2: 28–36. - PubMed
-
- Blakely, G. , Colloms S., May G., Burke M., and Sherratt D.. 1991. “ <styled-content style="fixed-case"> Escherichia coli </styled-content> XerC Recombinase Is Required for Chromosomal Segregation at Cell Division.” New Biologist 3, no. 8: 789–798. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

