Expression Profile of Serum CircFUNDC1 and CircUHRF1 Can Differentiate Between Colorectal Cancer and Inflammatory Bowel Diseases (Ulcerative Colitis and Crohn's Disease)
- PMID: 40376950
- PMCID: PMC12144590
- DOI: 10.1002/jcla.70039
Expression Profile of Serum CircFUNDC1 and CircUHRF1 Can Differentiate Between Colorectal Cancer and Inflammatory Bowel Diseases (Ulcerative Colitis and Crohn's Disease)
Abstract
Background: Colorectal cancer (CRC) is a worldwide burden. Circular RNAs are promising biomarkers for diagnosing and prognosis of CRC.
Objective: To investigate the possible association of sera levels of CircFUNDC1 and CircUHRF1 expression with predisposition and clinicopathological findings in CRC, ulcerative colitis (UC), and Crohn's disease (CD) in Egyptian patients.
Methods: The serum levels of CircFUNDC1 and CircUHRF1 were evaluated in 113 Egyptian subjects divided into four groups; CRC (31), UC (26), and CD (25) and compared to healthy controls (31) using quantitative polymerase chain reaction.
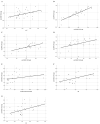
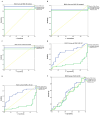
Results: The median values of log2 serum fold change (FC) of CircFUNDC1 in CRC, UC, and CD patients were 9.11, 6.58, and 6.17, respectively. It was upregulated in all case groups. CRC, UC, and CD patients had significantly higher serum CircFUNDC1 levels than controls (p < 0.001). However, there were no significant differences among patient groups (CRC, UC, and CD). The medians of log 2 of serum FC CircUHRF1 in patients with CRC, UC, and CD were -2.00, 3.33, and 3.12, respectively. The CircUHRF1 serum level was lower in the CRC group of patients, with no significant difference between the CRC group and the controls. Serum CircUHRF1 was significantly overexpressed in patients with UC and CD compared to the CRC groups or controls (p < 0.001). By Roc curve analysis, both genes can differentiate CRC patients from inflammatory bowel disease (IBD) patients or healthy controls with p < 0.05.
Conclusion: Serum CircFUNDC1 is a biomarker for CRC, while CircUHRF1 is a biomarker of IBD.
Keywords: CircFUNDC1; CircUHRF1; Colorectal cancer; Crohn's disease; Ulcerative colitis.
© 2025 The Author(s). Journal of Clinical Laboratory Analysis published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

