Glucose, glutamine, lactic acid and α‑ketoglutarate restore metabolic disturbances and atrophic changes in energy‑deprived muscle cells
- PMID: 40376969
- PMCID: PMC12105454
- DOI: 10.3892/mmr.2025.13562
Glucose, glutamine, lactic acid and α‑ketoglutarate restore metabolic disturbances and atrophic changes in energy‑deprived muscle cells
Abstract
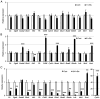
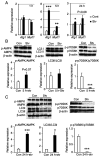
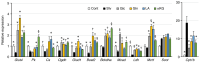
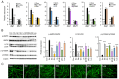
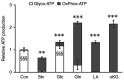
Skeletal muscle atrophy is often triggered by catabolic conditions such as fasting, malnutrition and chronic diseases; however, the efficacy of nutritional supplementation in maintaining muscle mass and preventing muscle atrophy remains controversial. The present study aimed to compare the inhibitory effects of various nutritional substrates on starvation‑induced catabolic changes and muscle cell atrophy. C2C12 muscle cells were starved for up to 24 h in medium lacking serum and main nutrients (glucose, glutamine and pyruvate). To assess the effects of exogenous substrates, the cells were incubated in starvation medium and individually supplemented with each of the following nutrients: Glucose, amino acids, fatty acids, lactate or ketone bodies. The expression of each gene and protein was analyzed by reverse transcription‑quantitative PCR and western blotting, respectively. Mitochondrial activity was determined by MTT assay and cell morphology was observed by immunofluorescence staining. The results revealed that starvation for >3 h suppressed mitochondrial activity, and after 5 h of starvation, the expression levels of several metabolic genes were increased; however, the levels of most, with the exception of Scot and Cpt‑1b, were suppressed after 24 h. Protein degradation and a decrease in protein synthesis were observed after 5 h of starvation, followed by autophagy with morphological atrophy at 24 h. Supplementation with specific substrates, with the exception of leucine, such as glucose, glutamine, lactic acid or α‑ketoglutarate, attenuated the suppression of mitochondrial activity, and altered gene expression, protein degradation and myotube atrophy in starved myotubes. Furthermore, the decrease in intracellular ATP production after 24 h of starvation was reversed by restoring glycolysis in glucose‑treated cells, and via an increase in mitochondrial respiration in cells treated with glutamine, lactic acid or α‑ketoglutarate. In conclusion, increasing the availability of glucose, glutamine, lactic acid or α‑ketoglutarate may be beneficial for countering muscle atrophy associated with inadequate nutrient intake.
Keywords: atrophy; metabolism; muscle cells; nutrient substrates; starvation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

