Shielding Human Adipocytes From Inflammation: The Protective Potential of Polyphenol-Rich Opuntia ficus-indica Cladode Extract
- PMID: 40377300
- PMCID: PMC12280845
- DOI: 10.1002/mnfr.70114
Shielding Human Adipocytes From Inflammation: The Protective Potential of Polyphenol-Rich Opuntia ficus-indica Cladode Extract
Abstract
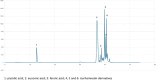
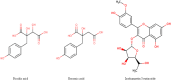
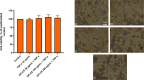
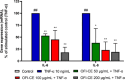
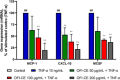
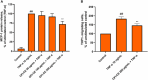
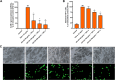
Opuntia ficus-indica (OFI) has attracted much attention as a source of antioxidant and antiinflammatory compounds. We hypothesize that the antioxidant content of OFI cladode extract may improve adipocyte dysfunction resulting from inflammatory stimulation of hypertrophic adipocytes. To this end, the properties of OFI cladode hydroalcoholic extract were evaluated in terms of antioxidant activity, regulation of adipocyte inflammation, and adipocyte/monocyte interaction in human adipocytes rendered dysfunctional by the proinflammatory cytokine tumor necrosis factor-α (TNF-α). The major phenolic compounds identified were isorhamnetin derivatives and phenolic acids, including piscidic and eucomic acids. Our results show that OFI cladode extract exhibits antiradical activities and reduces the adhesion and transmigration activity of monocytes to inflamed adipocytes by inhibiting various cytokines, chemokines, and adhesion molecules such as interleukin (IL)-6 and IL-8 by ∼80%, monocyte chemotactic protein (MCP)-1, C-X-C motif chemokine ligand (CXC-L)10, macrophage colony-stimulating factor (M-CSF) from 40% to 50%, and intercellular adhesion molecule-1 (ICAM-1) by 70% at the higher concentration. In structurally and mechanistically by protein-ligand docking profiling study, piscidic acid proved to be the best potential candidate for a regulatory interaction with the activities of nuclear factor erythroid 2-related factor 2 (NRF-2) and nuclear factor-κB (NF-κB). In summary, these data highlight the potential of OFI as a dietary supplement in nutritional treatments aimed at combating the inflammatory stigmata of obesity.
Keywords: Opuntia ficus‐indica (L.) Mill; adipocytes; eucomic acids; inflammation; isorhamnetin; piscidic.
© 2025 The Author(s). Molecular Nutrition & Food Research published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- World Health Organization (2021) Obesity and overweight, https://www.who.int/news‐room/fact‐sheets/detail/obesity‐and‐overweight.
MeSH terms
Substances
Grants and funding
- Next Generation EU
- PE00000003/National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3-Call for proposals No. 341 of 15 March 2022 of Italian Ministry of University and Research funded by the European Union-Next Generation EU
- PRIN-2022 Prot. 2022NZNZH8/Italian Ministry of University and Research
- PRIN 2022 prot. 2022CAKEW9/Italian Ministry of University and Research
- CUP D93C22000890001/Italian Ministry of University and Research
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

