Crp and Arc system directly regulate the transcription of NADH dehydrogenase genes in Shewanella oneidensis nitrate and nitrite respiration
- PMID: 40377311
- PMCID: PMC12210985
- DOI: 10.1128/spectrum.03324-24
Crp and Arc system directly regulate the transcription of NADH dehydrogenase genes in Shewanella oneidensis nitrate and nitrite respiration
Abstract
NADH oxidation by NADH dehydrogenases (NDHs) is crucial for feeding respiratory quinone pool and maintaining the balance of NADH/NAD+. In the respiratory model organism Shewanella oneidensis, which possesses four NDHs, the longstanding notion had been that NDHs were not required under anoxic conditions until recent studies demonstrated their role in extracellular electron transfer. However, the role of each NDH, particularly under anoxic conditions, has not been characterized. Here, we systematically investigated the role of each NDH in aerobic and anaerobic nitrate and nitrite respiration using NDH triple mutants. We corroborated the involvement of NDHs in anaerobic nitrate/nitrite respiration, revealing different repertoires of NDHs employed by S. oneidensis in response to electron acceptor (EA) availability. The transcript levels of two nqrs were modulated by the EA conversion from nitrate to nitrite. Furthermore, we demonstrated that the global regulators Crp and the Arc system both directly controlled the transcription of four NDHs during nitrate/nitrite respiration. This study confirms the requirement of NDHs for anaerobic nitrate and nitrite respiration and sheds light on the respiratory remodeling mechanism whereby global regulators coordinate NDH genes transcription to adapt to redox-stratified environments.IMPORTANCENADH is an important electron source for the respiratory quinone pool. Multiple NADH dehydrogenases (NDHs) are widely present in prokaryotes, indicating the flexibility in NADH oxidation. As a renowned respiratory versatile model strain, Shewanella oneidensis possesses four NDHs, encompassing all three types of NDHs, with varying ion-translocating efficiencies. The redundancy of NDHs may confer advantages for S. oneidensis to survive and thrive in redox-stratified environments. However, the roles of each NDH, especially in anaerobic respiration, are less understood. Here, we evaluated the role of each NDH in aerobic and anaerobic nitrate/nitrite respiration. We found that the conversion of electron acceptor from nitrate to nitrite triggered the changes in the transcriptional levels of NDH genes, and global regulators Crp and the Arc system were involved in these processes. These findings elucidate the mechanism of the respiratory chain remodeling at the NADH oxidation step in response to different electron acceptors.
Keywords: NADH dehydrogenases; Shewanella oneidensis; global regulator; respiratory chain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



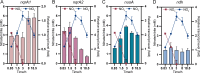
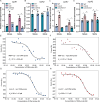

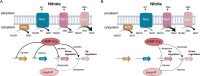
References
-
- Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

