Distinct region-specific neutralization profiles of contemporary HIV-1 clade C against best-in-class broadly neutralizing antibodies
- PMID: 40377318
- PMCID: PMC7617755
- DOI: 10.1128/jvi.00008-25
Distinct region-specific neutralization profiles of contemporary HIV-1 clade C against best-in-class broadly neutralizing antibodies
Abstract
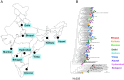
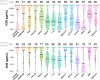
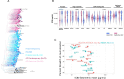
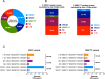
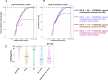
While broadly neutralizing antibodies (bnAbs) have been clinically shown to prevent HIV-1 acquisition, their relative effectiveness against regionally relevant HIV-1 forms is not clear. In the present study, we examined the extent of neutralization susceptibility of contemporary HIV-1 Indian clade C at a population level along with a head-to-head comparison with that from South Africa against a panel of clinically relevant best-in-class bnAbs. Env-pseudotyped viruses encoding HIV-1 India clade C env were found to be best neutralized by the V3 glycan-directed bnAbs (10-1074 and BG18) and select CD4 binding site (CD4bs)-directed bnAbs (VRC07, N6, and 1-18); however, they demonstrated significant resistance to V1/V2 apex-directed bnAbs. Interestingly, the magnitude of the neutralization sensitivity differed between contemporary India and South Africa clade C. Neutralization resistance to key bnAbs was observed to be associated with differences in residues on Env that form bnAb contact sites, gp120 loop lengths, and potential N-linked glycans. Notably, the second generation CD4bs bnAbs (VRC07, N6, 1-18) showed neutralization of VRC01- and 3BNC117-resistant viruses but with two- to sevenfold reduced potency compared to the VRC01-sensitive counterparts, likely due to the enrichment of resistance-associated residues observed in loop D. Predictive analysis indicated that the combination of BG18, N6, and PGDM1400 can provide over 95% neutralization coverage of contemporary India clade C at 1 µg/mL (IC80), an observation distinct from that observed with Africa clade C. Our study clearly highlights that both the complementarity of bnAb classes and the regionally relevant HIV-1 forms are important in achieving clinical effectiveness.IMPORTANCEWhile the development of vaccines to prevent HIV infection remains a global priority, their potential effectiveness is limited by the extraordinarily diversified circulating forms of HIV-1. The prospect of best-in-class broadly neutralizing antibodies (bnAbs) as a potential prevention option has been demonstrated in several studies, including the phase 2b Antibody-Mediated Prevention trials; however, to be broadly applicable, bnAbs will need to overcome the substantial variability of HIV env circulating globally, beyond the regions where efficacy trials are conducted. The present study highlights that the region-specific contemporary HIV-1 clade C viruses not only vary in their degree of susceptibility to the best-in-class clinically relevant bnAbs, but also are evolving at a population level to become increasingly resistant to the best-in-class bnAbs. Overall, the outcome of this study highlights the need for periodic assessment of sequence and neutralization profiles of the circulating regionally relevant HIV-1 forms toward prioritizing the bnAb combination suitable for effective intervention.
Keywords: HIV-1; India; South Africa; bnAb; clade C; contemporary virus; envelope; genetic diversity; neutralizing antibodies; prevention.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- UNAIDS . 2023. UNAIDS Global AIDS Update
-
- Carter A, Zhang M, Tram KH, Walters MK, Jahagirdar D, Brewer ED, Novotney A, Lasher D, Mpolya EA, Vongpradith A, et al. 2024. Global, regional, and national burden of HIV/AIDS, 1990-2021, and forecasts to 2050, for 204 countries and territories: the Global Burden of Disease Study 2021. Lancet HIV 11:e807–e822. doi: 10.1016/S2352-3018(24)00212-1 - DOI - PMC - PubMed
-
- Bertagnolio S, Hermans L, Jordan MR, Avila-Rios S, Iwuji C, Derache A, Delaporte E, Wensing A, Aves T, Borhan ASM, Leenus A, Parkin N, Doherty M, Inzaule S, Mbuagbaw L. 2021. Clinical impact of pretreatment human immunodeficiency virus drug resistance in people initiating nonnucleoside reverse transcriptase inhibitor-containing antiretroviral therapy: a systematic review and meta-analysis. J Infect Dis 224:377–388. doi: 10.1093/infdis/jiaa683 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- IA/TSG/19/1/600019/WTDBT_/DBT-Wellcome Trust India Alliance/India
- IA/TSG/19/1/600019/Wellcome Trust DBT India Alliance (India Alliance)
- BT/PR39156/DRUG/134/91/2021/Department of Biotechnology, Ministry of Science and Technology, India
- ADVANCE/USAID/International AIDS Vaccine Initiative
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

