Recurrent and Novel Pathogenic Variants in Genes Involved with Hearing Loss in the Pakistani Population
- PMID: 40377830
- PMCID: PMC12227476
- DOI: 10.1007/s40291-025-00782-w
Recurrent and Novel Pathogenic Variants in Genes Involved with Hearing Loss in the Pakistani Population
Abstract
Background: Molecular diagnostic rates for hereditary hearing loss vary by genetic ancestry, highlighting the importance of population-specific studies. In Pakistan, where consanguineous marriages are prevalent, genetic research has identified many autosomal recessive genes, advancing understanding of rare and novel hearing loss mechanisms. This study aimed to identify pathogenic genetic variants in 31 families from Azad Kashmir, Pakistan, presenting non-syndromic hearing loss.
Methods: We conducted exome sequencing and bioinformatics analysis, and targeted gene sequencing on 31 Pakistani families with hearing loss.
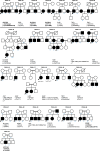
Results: We identified ten pathogenic, three likely pathogenic variants, and one variant of uncertain significance, comprising six nonsense, four missense, three frameshift, and one deep intronic variant, across ten hearing loss-associated genes (MYO15A, GJB2, SLC26A4, TMC1, HGF, TMIE, SLC19A2, KCNE1, ILDR, PCDH15 and MYO6) in 25 families. The overall diagnostic rate, including families with pathogenic and likely pathogenic variants, was 77.4%. GJB2 was the most frequently affected gene, identified in seven families. Thirteen out of 14 identified variants were homozygous. Notably, we identified two novel variants: MYO15A (NM_016239.4, DFNB3) c.870C>G, p.(Tyr290*) and MYO6 (NM_016239.4, DFNB37) c.3465del, p.(Pro1156Leufs*9). Additionally, we identified c.10475dupA, p.(Leu3493Alafs*25) in MYO15A (NM_016239.4, DFNB3) and c.617T>A, p.(Leu206*) in SLC26A4 (NM_000441.2, DFNB4), previously documented in ClinVar but unpublished. We also propose SLC19A2 as a candidate gene presenting as non-syndromic hearing loss, despite its association with thiamine-responsive megaloblastic anemia syndrome.
Conclusion: Our work expands the genotypic and phenotypic spectrum of hearing loss by emphasizing the importance of investigating under-represented groups to identify unique genetic variants and clinical characteristics. Such efforts deepen understanding of genetic diversity in under-represented populations to improve diagnosis and treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Funding: Open Access funding enabled and organized by Projekt DEAL. This work was supported by the German Research Foundation (DFG) VO 2138/7-1 grant 469177153, the DFG Heisenberg program VO 2138/8-1 grant 543719215, the DFG Collaborative Research Center 1690 (Project A03 to BV), and the Higher Education Commission (HEC) Pakistan under IRSIP Fellowship Program. Conflict of interest: M.S., A.B.M., L.N.M.V., A.A.A., B.K., A.E., Z.L., V.G., D.O., M.H.J., Z.Z., R.M., H.H., H.G.K., and B.V. have no conflicts of interest. All authors have read and agreed to the published version of the manuscript. Ethics approval: This study was approved by the Institutional Review board of Mirpur University of Science and Technology, Mirpur AJK, Pakistan (approval number NO.ORIC/301/2023 dated 11-09-2023). Consent to participate: Informed consent was obtained from all participants in the study, or from parents/legal guardians in the case of minors. Consent for publication: After reviewing participant consent letters for publishing, the Institutional Review board of Mirpur University of Science and Technology, Mirpur AJK, Pakistan granted consent. Availability of data and material: The data supporting the findings of this study are available from the corresponding author, B.V., upon reasonable request. Variants have been submitted to ClinVar under SUB14945752. Code availability: Not applicable. Author contributions: Conceptualization, M.S.; methodology, and software, B.V., M.S., L.M., A.E., A.B.M., R.M., B.K.,V.G., M.H.J., L.N.M.V. validation, B.V., D.O., Z.Z., L.N.M.V. writing—original draft preparation, M.S. writing—review and editing visualization, A.A.A., B.V., H.G.K., M.S. and supervision, A.A.A., Z.L., H.G.K., H.H., and B.V. All authors have read and agreed to the published version of the article.
Figures



References
-
- Zuriekat M, Qarmout S, Alsous M, Nanah A, Al-Halasa F, Alqudah S, et al. Hearing loss in Jordan: an overlooked public health challenge: Hearing loss and hearing healthcare in Jordan. Jordan Med J. 2024;58.
-
- Shave S, Botti C, Kwong K. Congenital sensorineural hearing loss. Pediatr Clin N Am. 2022;69:221–34. - PubMed
-
- Hussain R, Bittles AH. The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci. 1998;30:261–75. - PubMed
-
- Maheen P, Saeed A, Maira A, Shabbir AM, Saher NU, Afshan A. Neonatal hearing screening program and challenges faced by the developing country: a tertiary care hospital experience. J Pak Med Assoc. 2023;1–15. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

