NLRP3 Inflammasome Activation Expands the Immunosuppressive Myeloid Stroma and Antagonizes the Therapeutic Benefit of STING Activation in Glioblastoma
- PMID: 40377974
- PMCID: PMC12163576
- DOI: 10.1158/2767-9764.CRC-23-0189
NLRP3 Inflammasome Activation Expands the Immunosuppressive Myeloid Stroma and Antagonizes the Therapeutic Benefit of STING Activation in Glioblastoma
Abstract
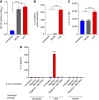
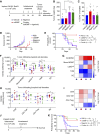
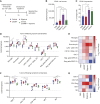
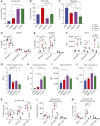
Glioblastoma (GBM) is the most common and deadly primary brain malignancy and is clinically refractory to immunotherapy. Active NLRP3 inflammasome signaling and IL-1β secretion have been observed in GBM, and NLRP3-driven myeloid-derived suppressor cell (MDSC) recruitment can mediate cancer immune evasion. Agonists of the cytosolic double-stranded DNA-sensing stimulator of IFN gene (STING) pathway can mediate proinflammatory conversion of cancer MDSCs; however, secretion of the NLRP3 products IL-1β and IL-18 has also been observed in certain myeloid populations following STING activation. In this study, we aimed to determine both the potential mechanistic synergy between STING and NLRP3 agonists, and the effects of this innate immune combination on the GBM tumor immune landscape. We find that STING activation does not prime pro-IL-1β expression for activated NLRP3 inflammasome secretion. In subcutaneous GL261 GBM, we show that NLRP3 activation expands the immunosuppressive myeloid stroma primarily via granulocytic MDSC recruitment and antagonizes the benefit of STING activation. In brain GL261, we find that NLRP3 activation expands granulocytic MDSCs but does not antagonize the therapeutic benefit of STING activation. Finally, we report that mesenchymal subtype GBM tumors have elevated neutrophil, IL-1β, and NLRP3 gene expression, a setting where our data suggest that NLRP3 activation could counteract STING agonists.
Significance: NLRP3 inflammasome signaling, which suppresses antitumor immunity in some cancers, has been observed in GBM tissues. NLRP3 activation in GBM induces granulocyte-dependent tumor immunosuppression and antagonizes the therapeutic efficacy of STING activation.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
S.T. Lea reports grants from The Joan Traver Walsh Foundation, The Brockman Foundation, the NIH National Center for Advancing Translational Sciences, and Cancer Prevention and Research Institute of Texas during the conduct of the study. C.-H. Chen reports grants from The Brockman Foundation and The Joan Traver Walsh Family Foundation during the conduct of the study. I. Lopez Del Castillo reports grants from The Joan Traver Walsh Foundation and The Brockman Foundation during the conduct of the study. M.A. Curran reports grants and personal fees from ImmunoGenesis, Inc. outside the submitted work, as well as a patent to Cyclic Dinucleotides as Agonists of Stimulator of Interferon Gene Dependent Signaling issued and licensed. No other disclosures were reported.
Figures

References
-
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013;310:1842–50. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

