Entabolons: How Metabolites Modify the Biochemical Function of Proteins and Cause the Correlated Behavior of Proteins in Pathways
- PMID: 40378093
- PMCID: PMC12152931
- DOI: 10.1021/acs.jcim.5c00462
Entabolons: How Metabolites Modify the Biochemical Function of Proteins and Cause the Correlated Behavior of Proteins in Pathways
Abstract
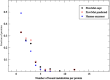
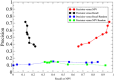
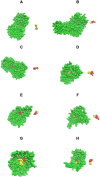
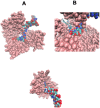
Although there are over 100,000 distinct human metabolites, their biological significance is often not fully appreciated. Metabolites can reshape the protein pockets to which they bind by COLIG formation, thereby influencing enzyme kinetics and altering the monomer-multimer equilibrium in protein complexes. Binding a common metabolite to a set of protein monomers or multimers results in metabolic entanglements that couple the conformational states and functions of nonhomologous, nonphysically interacting proteins that bind the same metabolite. These shared metabolites might provide the collective behavior responsible for protein pathway formation. Proteins whose binding and functional behavior is modified by a set of metabolites are termed an "entabolon"─a portmanteau of metabolic entanglement and metabolon. 55%-60% (22%-24%) of pairs of nonenzymatic proteins that likely bind the same metabolite have a p-value that they are in the same pathway, which is <0.05 (0.0005). Interestingly, the most populated pairs of proteins common to multiple pathways bind ancient metabolites. Similarly, we suggest how metabolites can possibly activate, terminate, or preclude transcription and other nucleic acid functions and may facilitate or inhibit the binding of nucleic acids to proteins, thereby influencing transcription and translation processes. Consequently, metabolites likely play a critical role in the organization and function of biological systems.
Figures




Similar articles
-
Implications of the Essential Role of Small Molecule Ligand Binding Pockets in Protein-Protein Interactions.J Phys Chem B. 2022 Sep 15;126(36):6853-6867. doi: 10.1021/acs.jpcb.2c04525. Epub 2022 Aug 31. J Phys Chem B. 2022. PMID: 36044742 Free PMC article.
-
Metabolite coupling in genome-scale metabolic networks.BMC Bioinformatics. 2006 Mar 6;7:111. doi: 10.1186/1471-2105-7-111. BMC Bioinformatics. 2006. PMID: 16519800 Free PMC article.
-
Studies of metabolite-protein interactions: a review.J Chromatogr B Analyt Technol Biomed Life Sci. 2014 Sep 1;966:48-58. doi: 10.1016/j.jchromb.2013.11.043. Epub 2013 Nov 25. J Chromatogr B Analyt Technol Biomed Life Sci. 2014. PMID: 24321277 Free PMC article. Review.
-
NMR identification of endogenous metabolites interacting with fatted and non-fatted human serum albumin in blood plasma: Fatty acids influence the HSA-metabolite interaction.J Magn Reson. 2013 Mar;228:81-94. doi: 10.1016/j.jmr.2012.12.010. Epub 2013 Jan 8. J Magn Reson. 2013. PMID: 23357430
-
Regulatory roles of metabolites in cell signaling networks.J Genet Genomics. 2013 Jul 20;40(7):367-74. doi: 10.1016/j.jgg.2013.05.002. Epub 2013 May 31. J Genet Genomics. 2013. PMID: 23876777 Review.
References
-
- Bionumbers What are the concentrations of free metabolites in cells?. 2024. https://book.bionumbers.org/what-are-the-concentrations-of-free-metaboli....
-
- Goodman, L. S. ; Brunton, L. L. ; Chabner, B. ; Knollmann, B. R. C. . Goodman & Gilman’s pharmacological basis of therapeutics; McGraw-Hill, 2011.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

