A large collection of bioinformatics question-query pairs over federated knowledge graphs: methodology and applications
- PMID: 40378136
- PMCID: PMC12083453
- DOI: 10.1093/gigascience/giaf045
A large collection of bioinformatics question-query pairs over federated knowledge graphs: methodology and applications
Abstract
Background: In recent decades, several life science resources have structured data using the same framework and made these accessible using the same query language to facilitate interoperability. Knowledge graphs have seen increased adoption in bioinformatics due to their advantages for representing data in a generic graph format. For example, yummydata.org catalogs more than 60 knowledge graphs accessible through SPARQL, a technical query language. Although SPARQL allows powerful, expressive queries, even across physically distributed knowledge graphs, formulating such queries is a challenge for most users. Therefore, to guide users in retrieving the relevant data, many of these resources provide representative examples. These examples can also be an important source of information for machine learning (for example, machine-learning algorithms for translating natural language questions to SPARQL), if a sufficiently large number of examples are provided and published in a common, machine-readable, and standardized format across different resources.
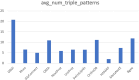
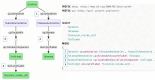
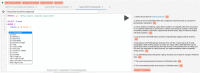
Findings: We introduce a large collection of human-written natural language questions and their corresponding SPARQL queries over federated bioinformatics knowledge graphs (KGs) collected for several years across different research groups at the SIB Swiss Institute of Bioinformatics. The collection comprises more than 1,000 example questions and queries, including almost 100 federated queries. We propose a methodology to uniformly represent the examples with minimal metadata, based on existing standards. Furthermore, we introduce an extensive set of open-source applications, including query graph visualizations and smart query editors, easily reusable by KG maintainers who adopt the proposed methodology.
Conclusions: We encourage the community to adopt and extend the proposed methodology, towards richer KG metadata and improved Semantic Web services. URL: https://github.com/sib-swiss/sparql-examples.
Keywords: Resource Description Framework (RDF); federated SPARQL; knowledge graphs; metadata; query editor.
© The Author(s) 2025. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Vollmers D, Srivastava N, Zahera HN. et al. UniQ-gen: unified query generation across multiple knowledge graphs. Knowledge Engineering and Knowledge Management. EKAW 2024. Lecture Notes in Computer Science, 2025. 10.1007/978-3-031-77792-9_11. - DOI
-
- Expasy overview of SIB Knowledge Graphs. [cited 20 Feb 2025]. https://www.expasy.org/search/SPARQL. Accessed 8 May 2025.
-
- Vrandečić D, Krötzsch M. Wikidata: a free collaborative knowledgebase. Commun ACM. 2014;57:78–85. 10.1145/2629489. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

