Hsc70-4: An unanticipated mediator of dsRNA internalization in Drosophila
- PMID: 40378201
- PMCID: PMC12083535
- DOI: 10.1126/sciadv.adv1286
Hsc70-4: An unanticipated mediator of dsRNA internalization in Drosophila
Abstract
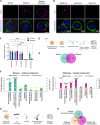
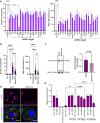
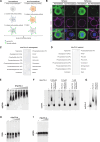
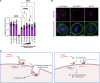
The small interfering RNA pathway is the primary antiviral defense mechanism in invertebrates and plants. This systemic mechanism relies on the recognition, transport, and internalization of double-stranded RNA (dsRNA). Our aim was to identify cell surface proteins that bind extracellular dsRNA and mediate its internalization in Drosophila cells. We used coimmunoprecipitation coupled with proteomics analysis and found that silencing heat shock cognate protein 70-4 (Hsc70-4), a constitutively expressed heat shock protein, impairs dsRNA internalization. Unexpectedly, despite lacking a predicted transmembrane domain, Hsc70-4 localizes to the cell membrane via lipid interactions. Antibody blocking experiments revealed an extracellular domain on Hsc70-4 that is essential for dsRNA internalization. Intriguingly, this dsRNA-specific binding capacity of Hsc70-4 functions independently of its chaperone activity. These findings not only highlight Hsc70-4 as a previously uncharacterized and essential component in the dsRNA internalization process but also offer promising insights for advancing RNA interference-based technologies to combat pests and vector-borne diseases.
Figures

References
-
- Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A., The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874 (2007). - PubMed
-
- Grant M., Lamb C., Systemic immunity. Curr. Opin. Plant Biol. 9, 414–420 (2006). - PubMed
-
- Karlikow M., Goic B., Saleh M. C., RNAi and antiviral defense in Drosophila: Setting up a systemic immune response. Dev. Comp. Immunol. 42, 85–92 (2014). - PubMed
-
- Kemp C., Mueller S., Goto A., Barbier V., Paro S., Bonnay F., Dostert C., Troxler L., Hetru C., Meignin C., Pfeffer S., Hoffmann J. A., Imler J.-L., Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658 (2013). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

