Native Top-Down Analysis of Membrane Protein Complexes Directly From In Vitro and Native Membranes
- PMID: 40378922
- PMCID: PMC12305242
- DOI: 10.1016/j.mcpro.2025.100993
Native Top-Down Analysis of Membrane Protein Complexes Directly From In Vitro and Native Membranes
Abstract
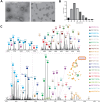
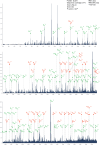
Macromolecular organization of proteins and lipids in cellular membranes is fundamental to cell functionality. Recent advances in native mass spectrometry (nMS) have established it as a key analytical tool for capturing these associations. This typically necessitates the extraction of target membrane proteins (MPs) from their physiological environments into detergent-like surroundings. In our recent studies using in vitro synthetic liposomes, we discovered that gas phase supercharging can selectively destabilize lipid bilayers and enable MS1 detection of embedded and associated protein-lipid complexes. Here, we further extend and apply this methodology to native cell-derived membrane vesicles. We demonstrate our ability to detect and ID protein complexes and their proteoforms directly from native membranes using supercharger-assisted prequadrupole activation followed by downstream native top-down tandem mass spectrometry, which combines both collision-based and electron capture-based fragmentation approaches. We first demonstrated this approach through native top-down identification of several integral MPs from in vitro membranes. Subsequently, we developed a protocol to produce nMS-ready native membrane vesicles. Applying to Escherichia coli total membranes, we generated nMS-ready vesicles and identified both integral and membrane-associated protein complexes of homomeric and heteromeric nature using our supercharging-enabled native top-down platform. For the heteropentameric β-barrel-assembly machinery (BAM) complex, which includes the integral MP BAM-A, we detected several lipidated proteoforms. For peripheral homodimeric dihydrolipoyl dehydrogenase, we identified bound endogenous metabolite cofactors. Furthermore, using BAM complex, a crucial antibiotic target, we show how this platform could be utilized to study drug binding to MPs directly from their native membranes.
Keywords: drug binding; electron capture fragmentation; membrane protein complex; native top–down mass spectrometry; proteoform analysis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




References
-
- Andersen O.S., Koeppe R.E. Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

