ARV1 is a component of the enzyme initiating glycosylphosphatidylinositol biosynthesis
- PMID: 40378954
- PMCID: PMC12182360
- DOI: 10.1016/j.jbc.2025.110236
ARV1 is a component of the enzyme initiating glycosylphosphatidylinositol biosynthesis
Abstract
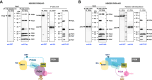
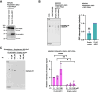
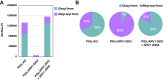
Glycosylphosphatidylinositol (GPI) serves as a membrane anchor of numerous cell surface proteins. It is synthesized in the endoplasmic reticulum from phosphatidylinositol (PI) by stepwise reactions and transferred to the C terminus of the protein. Defects in genes involved in GPI biosynthesis affect the expression of GPI-anchored proteins or their structure, causing the neurological disorder, inherited GPI deficiency. Individuals with ARV1 deficiency have symptoms resembling inherited GPI deficiency, but how ARV1 regulates GPI biosynthesis is poorly understood. Here, we show that ARV1 acts as a component of the enzyme initiating GPI biosynthesis, GPI N-acetylglucosaminyltransferase (GPI-GnT) complex, which forms a ring structure as predicted by AlphaFold3. ARV1 associates with PIGQ, a GPI-GnT component, and ARV1 mutants defective in this association lose their ability to enhance GPI-GnT activity, showing that association with PIGQ is critical for ARV1's function. ARV1-containing GPI-GnT used PI more efficiently than ARV1-less GPI-GnT in an in vitro enzyme assay. Collectively, our results suggest that ARV1 facilitates efficient recruitment of PI to GPI-GnT, thereby playing a critical role in the regulation of GPI-anchored protein expression.
Keywords: AlphaFold; GPI N-acetylglucosaminyltransferase complex; inherited GPI deficiency; lipidomics; phosphatidylinositol.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Komath S.S., Fujita M., Hart G.W., Ferguson M.A.J., Kinoshita T. In: Glycosylphosphatidylinositol Anchors in Essentials of Glycobiology. 4th Ed. Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., et al., editors. Cold Spring Harbor; Cold Spring Harbor, NY: 2022. pp. 141–154.
-
- Duval R., Nicolas G., Willemetz A., Murakami Y., Mikdar M., Vrignaud C., et al. Inherited glycosylphosphatidylinositol defects cause the rare Emm-negative blood phenotype and developmental disorders. Blood. 2021;137:3660–3669. - PubMed
-
- Miyata T., Takeda J., Iida Y., Yamada N., Inoue N., Takahashi M., et al. The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science. 1993;259:1318–1320. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

