NDRG1 and its family members: More than just metastasis suppressor proteins and targets of thiosemicarbazones
- PMID: 40378957
- PMCID: PMC12272907
- DOI: 10.1016/j.jbc.2025.110230
NDRG1 and its family members: More than just metastasis suppressor proteins and targets of thiosemicarbazones
Abstract
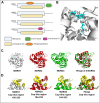
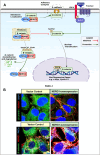
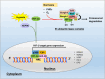
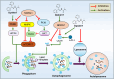
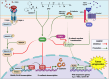
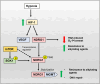
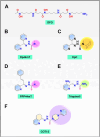
N-Myc downstream regulated gene-1 (NDRG1) and the other three members of this family (NDRG2, 3, and 4) play various functional roles in the cellular stress response, differentiation, migration, and development. These proteins are involved in regulating key signaling proteins and pathways that are often dysregulated in cancer, such as EGFR, PI3K/AKT, c-Met, and the Wnt pathway. NDRG1 is the primary, well-examined member of the NDRG family and is generally characterized as a metastasis suppressor that inhibits the first step in metastasis, the epithelial-mesenchymal transition. While NDRG1 is well-studied, emerging evidence suggests NDRG2, NDRG3, and NDRG4 also play significant roles in modulating oncogenic signaling and cellular homeostasis. NDRG family members are regulated by multiple mechanisms, including transcriptional control by hypoxia-inducible factors, p53, and Myc, as well as post-translational modifications such as phosphorylation, ubiquitination, and acetylation. Pharmacological targeting of the NDRG family is a therapeutic strategy against cancer. For instance, di-2-pyridylketone 4,4-dimethyl-3-thiosemicarbazone (Dp44mT) and di-2-pyridylketone-4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC) have been extensively shown to upregulate NDRG1 expression, leading to metastasis suppression and inhibition of tumor growth in multiple cancer models. Similarly, targeting NDRG2 demonstrates its pro-apoptotic and anti-proliferative effects, particularly in glioblastoma and colorectal cancer. This review provides a comprehensive analysis of the structural features, regulatory mechanisms, and biological functions of the NDRG family and their roles in cancer and neurodegenerative diseases. Additionally, NDRG1-4 are explored as therapeutic targets in oncology, focusing on recent advances in anti-cancer agents that induce the expression of these proteins. Implications for future research and clinical applications are also discussed.
Keywords: NDRG1; metastasis; metastasis suppression; thiosemicarbazone.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
References
-
- Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. - PubMed
-
- Bandyopadhyay S., Pai S.K., Gross S.C., Hirota S., Hosobe S., Miura K., et al. The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res. 2003;63:1731–1736. - PubMed
-
- Guan R.J., Ford H.L., Fu Y., Li Y., Shaw L.M., Pardee A.B. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res. 2000;60:749–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

