Throwing a spotlight on genomic dark matter: The power and potential of transposon-insertion sequencing
- PMID: 40378959
- PMCID: PMC12173739
- DOI: 10.1016/j.jbc.2025.110231
Throwing a spotlight on genomic dark matter: The power and potential of transposon-insertion sequencing
Abstract
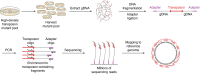
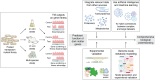
Linking genotype to phenotype is a central goal in biology. In the microbiological field, transposon mutagenesis is a technique that has been widely used since the 1970s to facilitate this connection. The development of modern 'omics approaches and next-generation sequencing have allowed high-throughput association between genes and their putative function. In 2009, four different variations in modern transposon-insertion sequencing (TIS) approaches were published, being referred to as transposon-directed insertion-site sequencing (TraDIS), transposon sequencing (Tn-seq), insertion sequencing (INSeq), and high-throughput insertion tracking by deep sequencing (HITS). These approaches exploit a similar concept to allow estimation of the essentiality or contribution to fitness of each gene in any bacterial genome. The main rationale is to perform a comparative analysis of the abundance of specific transposon mutants under one or more selective conditions. The approaches themselves only vary in the transposon used for mutagenesis, and in the methodology used for sequencing library preparation. In this review, we discuss how TIS approaches have been used to facilitate a major shift in our fundamental understanding of bacterial biology in a range of areas. We focus on several aspects including pathogenesis, biofilm development, polymicrobial interactions in various ecosystems, and antimicrobial resistance. These studies have provided new insight into bacterial physiology and revealed predicted functions for hundreds of genes previously representing genomic "dark matter." We also discuss how TIS approaches have been used to understand complex bacterial systems and interactions and how future developments of TIS could continue to accelerate and enrich our understanding of bacterial biology.
Keywords: Tn-seq; TraDIS; bacterial genomics; dark matter; high-throughput screening; linking genotype-phenotype; novel genes; transposon mutagenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. - PubMed
-
- Hunt M., Lima L., Shen W., Lees J., Iqbal Z. AllTheBacteria - all bacterial genomes assembled, available and searchable. BioRxiv. 2024 doi: 10.1101/2024.03.08.584059. - DOI
-
- O’Toole G.A., Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998;28:449–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

