Alternative splicing across the C. elegans nervous system
- PMID: 40379606
- PMCID: PMC12084653
- DOI: 10.1038/s41467-025-58293-5
Alternative splicing across the C. elegans nervous system
Abstract
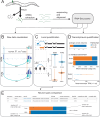
Alternative splicing is a key mechanism that shapes transcriptomes, helping to define neuronal identity and modulate function. Here, we present an atlas of alternative splicing across the nervous system of Caenorhabditis elegans. Our analysis identifies novel alternative splicing in key neuronal genes such as unc-40/DCC and sax-3/ROBO. Globally, we delineate patterns of differential alternative splicing in almost 2000 genes, and estimate that a quarter of neuronal genes undergo differential splicing. We introduce a web interface for examination of splicing patterns across neuron types. We explore the relationship between neuron type and splicing, and between splicing and differential gene expression. We identify RNA features that correlate with differential alternative splicing and describe the enrichment of microexons. Finally, we compute a splicing regulatory network that can be used to generate hypotheses on the regulation and targets of alternative splicing in neurons.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet40, 1413–1415 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

