ZSCAN21 mediates the pathogenic transcriptional induction of α-synuclein in cellular and animal models of Parkinson's disease
- PMID: 40379611
- PMCID: PMC12084422
- DOI: 10.1038/s41419-025-07722-w
ZSCAN21 mediates the pathogenic transcriptional induction of α-synuclein in cellular and animal models of Parkinson's disease
Abstract
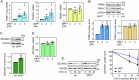
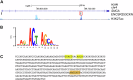
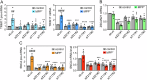
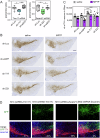
The expression level of α-synuclein is thought to play a crucial role in the pathogenesis of Parkinson's disease. However, little is known about the molecular mechanisms regulating the transcription of its gene, SNCA, particularly in the context of the disease. The transcription factor ZSCAN21 has been shown to act on SNCA, but whether ZSCAN21 is actually involved in the induction of SNCA transcription in Parkinson's disease is unknown. To address this question, we used the MPTP mouse model and LUHMES-derived dopaminergic neuronal spheroids, subjected to Parkinson's disease-related neurotoxins and mutations. We show that MPP+-treated spheroids recapitulate the main features of α-synuclein pathology and that MPP+-triggered transcriptional induction of SNCA is associated with ZSCAN21 stabilisation. Importantly, knock-down of ZSCAN21 prevents both the MPP+-triggered increase in α-synuclein mRNA and pre-mRNA levels in LUHMES-derived spheroids and the death of dopaminergic neurons in the substantia nigra of MPTP-treated mice. These effects are recapitulated by knockdown of TRIM17, a ZSCAN21 stabiliser which prevents its ubiquitination and degradation mediated by TRIM41. Moreover, reducing the interaction between ZSCAN21 and TRIM41, either by inserting Parkinson's disease-associated mutations into the TRIM41 gene or by preventing SUMOylation of ZSCAN21, results in both stabilisation of ZSCAN21 and induction of SNCA. Taken together, our data strongly suggest that ZSCAN21 is a crucial transcription factor for pathogenic α-synuclein expression and neurodegeneration in Parkinson's disease, pointing to its regulators, TRIM17 and TRIM41, as original therapeutic targets for a neuroprotective treatment of Parkinson's disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: All the experimental and surgical procedures were conducted in accordance with the European (Directive 2010/63/UE), Spanish (Real Decreto 53/2013) and Catalan (Decret 214/97) legislation on the protection of animals used for experimental and other scientific purposes, and approved by the Vall d’Hebron Research Institute (VHIR) Ethical Experimentation Committee to ensure the use of the minimum necessary number of animals and the application of protocols that cause the least pain, suffering or distress to animals.
Figures



References
-
- Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Prim. 2017;3:17013. - PubMed
-
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–7. - PubMed
-
- Mochizuki H, Choong CJ, Masliah E. A refined concept: alpha-synuclein dysregulation disease. Neurochem Int. 2018;119:84–96. - PubMed
MeSH terms
Substances
Grants and funding
- 813599/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- 813599/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- MND202310017917/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- MND202310017917/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- 12372/Michael J. Fox Foundation for Parkinson's Research (Michael J. Fox Foundation)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

