USP2 inhibition unleashes CD47-restrained phagocytosis and enhances anti-tumor immunity
- PMID: 40379682
- PMCID: PMC12084640
- DOI: 10.1038/s41467-025-59621-5
USP2 inhibition unleashes CD47-restrained phagocytosis and enhances anti-tumor immunity
Abstract
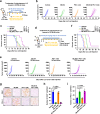
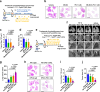
The CD47/SIRPα axis conveys a 'don't eat me' signal, thereby thwarting the phagocytic clearance of tumor cells. Although blocking antibodies targeting CD47 have demonstrated promising anti-tumor effects in preclinical models, clinical trials involving human cancer patients have not yielded ideal results. Exploring the regulatory mechanisms of CD47 is imperative for devising more efficacious combinational therapies. Here, we report that inhibiting USP2 prompts CD47 degradation and reshapes the tumor microenvironment (TME), thereby enhancing anti-PD-1 immunotherapy. Mechanistically, USP2 interacts with CD47, stabilizing it through deubiquitination. USP2 inhibition destabilizes CD47, thereby boosting macrophage phagocytosis. Single-cell RNA sequencing shows USP2 inhibition reprograms TME, evidenced by increasing M1 macrophages and CD8+ T cells while reducing M2 macrophages. Combining ML364 with anti-PD-1 reduces tumor burden in mouse models. Clinically, low USP2 expression predicts a better response to anti-PD-1 treatment. Our findings uncover the regulatory mechanism of CD47 by USP2 and targeting this axis boosts anti-tumor immunity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Bagchi, S., Yuan, R. & Engleman, E. G. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu. Rev. Pathol.16, 223–249 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

