Application of the joint clustering algorithm based on Gaussian kernels and differential privacy in lung cancer identification
- PMID: 40379735
- PMCID: PMC12084312
- DOI: 10.1038/s41598-025-01873-8
Application of the joint clustering algorithm based on Gaussian kernels and differential privacy in lung cancer identification
Abstract
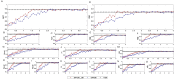
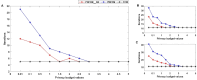
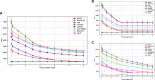
In the age of big data, privacy, particularly medical data privacy, is becoming increasingly important. Differential privacy (DP) has emerged as a key method for safeguarding privacy during data analysis and publishing. Cancer identification and classification play a vital role in early detection and treatment. This paper introduces a novel algorithm, DPFCM_GK, which combines differential privacy with fuzzy c-means (FCM) clustering using a Gaussian kernel function. The algorithm enhances cancer detection while ensuring data privacy. Three publicly available lung cancer datasets, along with a dataset from our hospital, are used to test and demonstrate the effectiveness of DPFCM_GK. The experimental results show that DPFCM_GK achieves high clustering accuracy and enhanced privacy as the privacy budget (ε) increases. For the UCIML, NLST, and NSCLC datasets, it reaches optimal results at lower ε (1.52, 1.24, and 2.32) compared to DPFCM. In the lung cancer dataset, DPFCM_GK outperforms DPFCM within, 0.05 ≤ ε ≤ 2.5, with significant differences (χ2 = 4.54 ∼ 29.12; P < 0.05), and both methods converge to an accuracy of 94.5% as ε increases. Although differential privacy initially increases iteration counts, DPFCM_GK demonstrates faster convergence and fewer iterations compared to DPFCM, with significant reductions (T= 23.08, 43.47, and 48.93; P<0.05). For the UCIML dataset, DPFCM_GK significantly reduces runtime compared to other models (DPFCM, LDP-SGD, LDP-Fed, LDP-FedSGD, MGM-DPL, LDP-FL) under the same privacy budget. The runtime reduction is statistically significant with T-values of (T = 21.08, 316.24, 102.35, 222.37, 162.23, 159.25; P < 0.05). DPFCM_GK still maintains excellent time efficiency when applied to the NLST and NSCLC datasets(P < 0.05). For the LLCS dataset, For the LLCS dataset, the DPFCM_GK demonstrates significant improvement as the privacy budget increases, especially in low-budget scenarios, where the performance gap is most pronounced (T=4.20, 8.44, 10.92, 3.95, 7.16, 8.51, P < 0.05). These results confirm DPFCM_GK as a practical solution for medical data analysis, balancing accuracy, privacy, and efficiency.
Keywords: Big data; DPFCM_GK; Differential privacy; Gaussian kernel function; Privacy budget; Privacy-preserving.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

