Dominant variants in major spliceosome U4 and U5 small nuclear RNA genes cause neurodevelopmental disorders through splicing disruption
- PMID: 40379786
- PMCID: PMC12165858
- DOI: 10.1038/s41588-025-02184-4
Dominant variants in major spliceosome U4 and U5 small nuclear RNA genes cause neurodevelopmental disorders through splicing disruption
Abstract
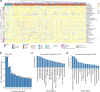
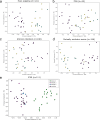
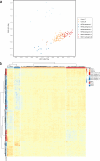
The major spliceosome contains five small nuclear RNAs (snRNAs; U1, U2, U4, U5 and U6) essential for splicing. Variants in RNU4-2, encoding U4, cause a neurodevelopmental disorder called ReNU syndrome. We investigated de novo variants in 50 snRNA-encoding genes in a French cohort of 23,649 individuals with rare disorders and gathered additional cases through international collaborations. Altogether, we identified 145 previously unreported probands with (likely) pathogenic variants in RNU4-2 and 21 individuals with de novo and/or recurrent variants in RNU5B-1 and RNU5A-1, encoding U5. Pathogenic variants typically arose de novo on the maternal allele and cluster in regions critical for splicing. RNU4-2 variants mainly localize to two structures, the stem III and T-loop/quasi-pseudoknot, which position the U6 ACAGAGA box for 5' splice site recognition and associate with different phenotypic severity. RNU4-2 variants result in specific defects in alternative 5' splice site usage and methylation patterns (episignatures) that correlate with variant location and clinical severity. This study establishes RNU5B-1 as a neurodevelopmental disorder gene, suggests RNU5A-1 as a strong candidate and highlights the role of de novo variants in snRNAs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.W. receives research funding from Novo Nordisk and has consulted for ArgoBio Studio. A.O’D.-L. is on the scientific advisory board for Congenica, was a paid consultant for Tome Biosciences and Ono Pharma USA, and at present for Addition Therapeutics, and received reagents from PacBio to support rare disease research. V. Jobanputra has served as a consultant to Illumina and received consulting fees from the company. All other authors declare no competing interests.
Figures














References
-
- Wilkinson, M. E., Charenton, C. & Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem.89, 359–388 (2020). - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

