APOBEC3 mutagenesis drives therapy resistance in breast cancer
- PMID: 40379787
- PMCID: PMC12165862
- DOI: 10.1038/s41588-025-02187-1
APOBEC3 mutagenesis drives therapy resistance in breast cancer
Abstract
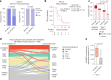
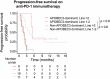
Acquired genetic alterations drive resistance to endocrine and targeted therapies in metastatic breast cancer; however, the underlying processes engendering these alterations are largely uncharacterized. To identify the underlying mutational processes, we utilized a clinically annotated cohort of 3,880 patient samples with tumor-normal sequencing. Mutational signatures associated with apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) enzymes were prevalent and enriched in post-treatment hormone receptor-positive cancers. These signatures correlated with shorter progression-free survival on antiestrogen plus CDK4/6 inhibitor therapy in hormone receptor-positive metastatic breast cancer. Whole-genome sequencing of breast cancer models and paired primary-metastatic samples demonstrated that active APOBEC3 mutagenesis promoted therapy resistance through characteristic alterations such as RB1 loss. Evidence of APOBEC3 activity in pretreatment samples illustrated its pervasive role in breast cancer evolution. These studies reveal APOBEC3 mutagenesis to be a frequent mediator of therapy resistance in breast cancer and highlight its potential as a biomarker and target for overcoming resistance.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.C. has received institutional grant/funding from Daiichi Sankyo, AstraZeneca, and Lilly, Shares/Ownership interests in Totus Medicines and consultation/Ad board/Honoraria from AstraZeneca, Lilly, Daiichi Sankyo, Novartis, Neogenomics, Nuvalent, Blueprint, SAGA Diagnostics and Effector Therapeutics. J.S.R.-F. is employed by AstraZeneca and reports receiving personal/consultancy fees from Goldman Sachs, Bain Capital, REPARE Therapeutics, Saga Diagnostics and Paige.AI, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas and ad hoc membership of the scientific advisory boards of AstraZeneca, Merck, Daiichi Sankyo, Roche Tissue Diagnostics and Personalis, outside the submitted work. P.R. has received institutional grant/funding from Grail, Novartis, AstraZeneca, Epic Sciences, Invitae/ArcherDx, Biothernostics, Tempus, Neogenomics, Biovica, Guardant, Personalis and Myriad, Shares/Ownership interests in Odyssey Biosciences and consultation/Ad board/Honoraria from Novartis, AstraZeneca, Pfizer, Lilly/Loxo, Prelude Therapeutics, Epic Sciences, Daiichi Sankyo, Foundation Medicine, Inivata, Natera, Tempus, SAGA Diagnostics, Paige.AI, Guardant and Myriad. S.N.P. has received funding for research from AstraZeneca, Philips and Varian, and reports consulting activity for Repare Therapeutics and AstraZeneca. N.R. has received research funding from BMS, Pfizer and REPARE therapeutics. B.W. reports research funding from Repare Therapeutics, outside the submitted work. F.P. is a member of the scientific advisory board of MultiplexDx. In addition, F.P. serves on the diagnostic advisory board and reports receiving consultancy fees from AstraZeneca. A. Gazzo reports consulting activity at enGenome and Janssen. The other authors declare no competing interests.
Figures











Update of
-
APOBEC3 mutagenesis drives therapy resistance in breast cancer.bioRxiv [Preprint]. 2024 May 1:2024.04.29.591453. doi: 10.1101/2024.04.29.591453. bioRxiv. 2024. Update in: Nat Genet. 2025 Jun;57(6):1452-1462. doi: 10.1038/s41588-025-02187-1. PMID: 38746158 Free PMC article. Updated. Preprint.
References
-
- Pearson, A. et al. Inactivating NF1 mutations are enriched in advanced breast cancer and contribute to endocrine therapy resistance. Clin. Cancer Res.26, 608–622 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- P01-CA234228/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P01 CA234228/CA/NCI NIH HHS/United States
- P50 CA247749/CA/NCI NIH HHS/United States
- P50-CA247749/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P50 CA247749 01/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

