MCSP+ metastasis founder cells activate immunosuppression early in human melanoma metastatic colonization
- PMID: 40379833
- PMCID: PMC12202500
- DOI: 10.1038/s43018-025-00963-w
MCSP+ metastasis founder cells activate immunosuppression early in human melanoma metastatic colonization
Abstract
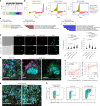
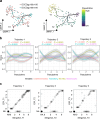
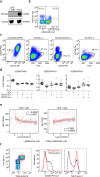
To investigate the early, poorly understood events driving metastatic progression, we searched for the earliest detectable disseminated cancer cells (DCCs), also often referred to as disseminated tumor cells (DTCs), in sentinel lymph node (SLN) biopsies of 492 patients with stage I-III melanoma. Using micromanipulator-assisted isolation of rare DCCs, single-cell mRNA and DNA sequencing, codetection by indexing immunofluorescence imaging and survival analysis, we identified melanoma-associated chondroitin sulfate proteoglycan (MCSP)+ melanoma cells as metastasis founder cells (MFCs). We found that DCCs entering SLNs predominantly exhibited a transitory phenotype that, upon interferon-γ exposure triggered by CD8 T cells, dedifferentiated into a neural-crest-like phenotype. This was accompanied by increased production of small extracellular vesicles (sEVs) carrying the immunomodulatory proteins CD155 and CD276 but rarely programmed cell death protein 1 ligand 1. The sEVs suppressed CD8 T cell proliferation and function, facilitating colony formation. Targeting MCSP+ MFCs or their immune escape mechanisms could be key to curing melanoma early by preventing manifestation of metastasis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures













References
MeSH terms
Substances
Grants and funding
- WE 4632/5-1//Deutsche Forschungsgemeinschaft (German Research Foundation)
- B09 FOR2127/Deutsche Forschungsgemeinschaft (German Research Foundation)
- B09/TRR 305/Deutsche Forschungsgemeinschaft (German Research Foundation)
- B13/TRR 305/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KL1233/14-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Z02/TRR 305/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KL1233/10-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KL1233/11-2/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 2015.016.1/Wilhelm Sander-Stiftung (Wilhelm Sander Foundation)
- 70112463/Deutsche Krebshilfe (German Cancer Aid)
- CAR-TIME, 101116768/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- INTERREG BY/CZ-118/EC | European Regional Development Fund (Europski Fond za Regionalni Razvoj)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

