Structural regulation of PLK1 activity: implications for cell cycle function and drug discovery
- PMID: 40379873
- PMCID: PMC12183085
- DOI: 10.1038/s41417-025-00907-7
Structural regulation of PLK1 activity: implications for cell cycle function and drug discovery
Abstract
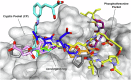
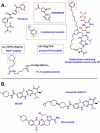
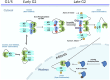
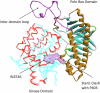
Polo Like Kinase 1 (PLK1), a key regulator of mitosis whose overexpression is often associated with poor survival rates in cancer, continues to be widely investigated as an oncology drug target with clinical trials evaluating second and third generation inhibitors. In addition to the conserved N-terminal kinase domain (KD), a unique characteristic of the Polo-Like kinase family is the C-terminal polo-box domain (PBD). The PBD contains a phosphopeptide binding site that recognizes substrates primed by other kinases and furthermore is responsible for subcellular localization of PLK1 to specific sites in the nucleus including centrosomes and kinetochores. Another role of the PBD is its regulatory ability through domain-domain interactions with the KD to maintain an autoinhibited state of PLK1. Insights into post translational modifications and the PBD - KD domain-domain association have been obtained and show that key events in PLK1 regulation include phosphosubstrate binding, T210 phosphorylation and engagement with the Bora protein. These can induce an open and active conformation where the domain-domain inhibitory interactions no longer dominate. Further regulatory events recently described include the interchange between monomeric and dimeric forms, which can also serve to inhibit or activate PLK1 during the cell cycle. Different oligomeric forms of PLK1, existing as homodimers and heterodimers with PLK2, have been identified and likely play context dependent roles. This review provides an overview of recent information describing structural and mechanistic insights into inhibition of PLK1 and the temporal and spatial requirements of its activation and regulation. It also covers recent insights into the conformational regulation of other members of the Polo-Like kinase family. The implications of the conformational regulation of PLK1 with respect to cell cycle function and drug discovery are significant and are therefore discussed in detail.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: In addition to his primary affiliation at the University of South Carolina, CM is President and CSO of PPI Pharmaceuticals, LLC, developing inhibitors as PLK1 as next generation cancer therapeutics. No other authors declare competing interests.
Figures




References
-
- Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nature Rev. 2010;9:643–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

