Atezolizumab plus bevacizumab and chemotherapy in metastatic nonsquamous NSCLC: the randomized double-blind phase 3 IMpower151 trial
- PMID: 40379995
- PMCID: PMC12283377
- DOI: 10.1038/s41591-025-03658-y
Atezolizumab plus bevacizumab and chemotherapy in metastatic nonsquamous NSCLC: the randomized double-blind phase 3 IMpower151 trial
Abstract
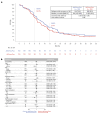
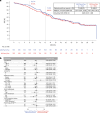
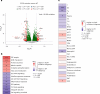
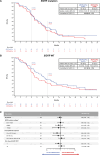
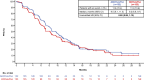
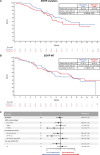
After the global approval of atezolizumab plus bevacizumab and chemotherapy as first-line metastatic nonsquamous non-small-cell lung cancer (nsqNSCLC) treatment, the IMpower151 ( NCT04194203 ) trial was conducted in China to address regional differences. Chemotherapy-naive patients with metastatic nsqNSCLC (N = 305) were randomized 1:1 to receive either atezolizumab, bevacizumab, carboplatin and paclitaxel or pemetrexed (ABCPem/Pac; n = 152) or placebo plus bevacizumab, carboplatin and pemetrexed or paclitaxel (BCPem/Pac; n = 153). The primary endpoint was investigator-assessed progression-free survival (INV-PFS); secondary endpoints included subgroup analyses of INV-PFS, independent review facility-assessed PFS, overall survival, and investigator-assessed objective response rate and duration of response per RECIST v.1.1. Most patients (97%) received pemetrexed, and 53% had EGFR+ tumors. Median INV-PFS for ABCPem/Pac versus BCPem/Pac was 9.5 versus 7.1 months (stratified hazard ratio: 0.84; 95% confidence interval: 0.65, 1.09; P = 0.184). INV-PFS across subgroups and independent review facility-assessed PFS were consistent with INV-PFS in the intention-to-treat population. Median overall survival was 20.7 versus 18.7 months in the ABCPem/Pac versus BCPem/Pac arms, respectively (stratified hazard ratio: 0.93; 95% confidence interval: 0.67, 1.28). Confirmed objective response rate with ABCPem/Pac versus BCPem/Pac was 48% versus 50%, respectively; median duration of response was 11.3 versus 8.3 months. Adverse events of special interest for atezolizumab were observed in 68% (grades 3 and 4: 11%) and 71% (grades 3 and 4: 7%) of patients receiving ABCPem/Pac and BCPem/Pac, respectively. The most common adverse events of special interest for atezolizumab in the ABCPem/Pac and BCPem/Pac arms were hepatitis (driven by laboratory abnormalities; mostly low grade), hypothyroidism and rash. Overall, IMpower151 did not meet its primary endpoint (INV-PFS) in metastatic nsqNSCLC. ABCPem/Pac was generally well tolerated, with no new safety signals. Trial registration number: ClinicalTrials.gov, NCT02366143.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.Z. received honoraria as a speaker/consultant from Alice, Amoy Diagnostics, AnHeart, Boehringer Ingelheim, C-Stone, Eli Lilly China, Hengrui, Innovent Biologics, LUYE Pharma, Merck Sharp & Dohme, Qilu, Roche, Sanofi and TopAlliance Biosciences Inc. along with advisor fees from Hengrui, Innovent Biologics, Qilu and TopAlliance Biosciences Inc. C.Z. declares non-financial competing interests as the IASLC President-Elect. I.Y., M.X., X.H., J. Cai and Q.Wu are employees of Roche (China) Holding Ltd and own stocks from Roche. L.Q. and C.S. are employees of Roche (China) Holding Ltd. M.B., M.K., M.K.S. are employees of Genentech and own stocks from Roche. The other authors declare no competing interests.
Figures


References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Socinski, M. A. et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N. Engl. J. Med.378, 2288–2301 (2018). - PubMed
-
- Reck, M. et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir. Med.7, 387–401 (2019). - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

