Genome-wide association analysis in peanut accessions uncovers the genetic basis regulating oil and fatty acid variation
- PMID: 40380082
- PMCID: PMC12082984
- DOI: 10.1186/s12870-025-06690-9
Genome-wide association analysis in peanut accessions uncovers the genetic basis regulating oil and fatty acid variation
Abstract
Background: The cultivated peanut, Arachis hypogaea L., is a critical oil and food crop worldwide. Improving seed oil quality in peanut has long been an aim of breeders. However, our knowledge of the genetic basis of selecting for seed nutritional traits is limited. Based on AhFAD2A and AhFAD2B, scientists have now developed higher oleic acid (80-84%) in peanut. Decoding the genetic makeup behind natural variation in kernel oil and fatty acid concentrations is crucial for molecular breeding-based nutrient quantity and quality manipulation.
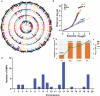
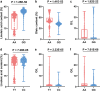
Results: Herein, we recognized 87 quantitative trait loci (QTLs) in 45 genomic regions for the concentrations of oil, oleic acid, and linoleic acid, as well as the oleic acid to linoleic acid (O/L) ratio via a genome-wide association study (GWAS) involving 499 peanut accessions. Eight QTLs explained more than 15% of the phenotypic variation in peanut accessions. Among the 45 potential genes significantly related to the four traits, only three genes displayed annotation to the fatty acid pathway. Furthermore, on the basis of pleiotropism or linkage data belonging to the identified singular QTLs, we generated a trait-locus axis to better elucidate the genetic background behind the observed oil and fatty acid concentration association. Expression analysis indicated that arahy.AV6GAN and arahy.NNA8KD have higher expressions in the seeds.
Conclusion: This natural population consisting of 499 peanut accessions combined with high-density SNPs will provide a better choice for identifying peanut QTLs/genes in the future. Together, our results provide strong evidence for the genetic mechanism behind oil biosynthesis in peanut, facilitating future advances in multiple fatty acid component generation via pyramiding of desirable QTLs.
Keywords: Fatty acid; GWAS; Genetic basis; Oil; Peanut.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.).BMC Genet. 2014 Dec 10;15:133. doi: 10.1186/s12863-014-0133-4. BMC Genet. 2014. PMID: 25491595 Free PMC article.
-
Genetic dissection of fatty acid components in the Chinese peanut (Arachis hypogaea L.) mini-core collection under multi-environments.PLoS One. 2022 Dec 30;17(12):e0279650. doi: 10.1371/journal.pone.0279650. eCollection 2022. PLoS One. 2022. PMID: 36584016 Free PMC article.
-
Genome-wide approaches delineate the additive, epistatic, and pleiotropic nature of variants controlling fatty acid composition in peanut (Arachis hypogaea L.).G3 (Bethesda). 2022 Jan 4;12(1):jkab382. doi: 10.1093/g3journal/jkab382. G3 (Bethesda). 2022. PMID: 34751378 Free PMC article.
-
Identification of potential QTLs and genes associated with seed composition traits in peanut (Arachis hypogaea L.) using GWAS and RNA-Seq analysis.Gene. 2021 Feb 15;769:145215. doi: 10.1016/j.gene.2020.145215. Epub 2020 Oct 7. Gene. 2021. PMID: 33038422
-
An Overview of Mapping Quantitative Trait Loci in Peanut (Arachis hypogaea L.).Genes (Basel). 2023 May 28;14(6):1176. doi: 10.3390/genes14061176. Genes (Basel). 2023. PMID: 37372356 Free PMC article. Review.
References
-
- Zhang L, Yang X, Tian L, Chen L, Yu W. Identification of peanut (Arachis hypogaea) chromosomes using a fluorescence in situ hybridization system reveals multiple hybridization events during tetraploid peanut formation. New Phytol. 2016;211(4):1424–39. - PubMed
-
- Isleib TG, Pattee HE, Sanders TH, Hendrix KW, Dean LO. Compositional and sensory comparisons between normal-and high-oleic peanuts. J Agric Food Chem. 2006;54(5):1759–63. - PubMed
MeSH terms
Substances
Grants and funding
- ZR2021QC172, ZR2023QC146/Natural Science Foundation of Shangdong Province
- ZR2021QC172, ZR2023QC146/Natural Science Foundation of Shangdong Province
- 2024LZGC035/Key R&D Program of Shandong Province
- KF2024007/Open Project of Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, P. R. China
- CXGC2023F20, CXGC2024F20, CXGC2024G20/the innovation Project of SAAS
- CXGC2023F20, CXGC2024F20, CXGC2024G20/the innovation Project of SAAS
- tstp20240523, tsqn202312292/Taishan Scholars Program
- tstp20240523, tsqn202312292/Taishan Scholars Program
- 2022E10012/Open Project of Key Laboratory of Digital Upland Crops of Zhejiang Province
- 2018GNC110036, 2022TZXD0031/Key research and development plan of Shandong Province
- 2018GNC110036, 2022TZXD0031/Key research and development plan of Shandong Province
- 2022A02008-3/Major scientific and technological project in Xinjiang
- CARS-13/China Agriculture Research System of MOF and MARA
LinkOut - more resources
Full Text Sources

