Deciphering microbial and metabolic influences in gastrointestinal diseases-unveiling their roles in gastric cancer, colorectal cancer, and inflammatory bowel disease
- PMID: 40380167
- PMCID: PMC12085054
- DOI: 10.1186/s12967-025-06552-w
Deciphering microbial and metabolic influences in gastrointestinal diseases-unveiling their roles in gastric cancer, colorectal cancer, and inflammatory bowel disease
Abstract
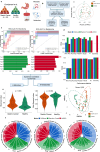
Introduction: Gastrointestinal disorders (GIDs) affect nearly 40% of the global population, with gut microbiome-metabolome interactions playing a crucial role in gastric cancer (GC), colorectal cancer (CRC), and inflammatory bowel disease (IBD). This study aims to investigate how microbial and metabolic alterations contribute to disease development and assess whether biomarkers identified in one disease could potentially be used to predict another, highlighting cross-disease applicability.
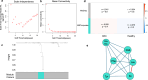
Methods: Microbiome and metabolome datasets from Erawijantari et al. (GC: n = 42, Healthy: n = 54), Franzosa et al. (IBD: n = 164, Healthy: n = 56), and Yachida et al. (CRC: n = 150, Healthy: n = 127) were subjected to three machine learning algorithms, eXtreme gradient boosting (XGBoost), Random Forest, and Least Absolute Shrinkage and Selection Operator (LASSO). Feature selection identified microbial and metabolite biomarkers unique to each disease and shared across conditions. A microbial community (MICOM) model simulated gut microbial growth and metabolite fluxes, revealing metabolic differences between healthy and diseased states. Finally, network analysis uncovered metabolite clusters associated with disease traits.
Results: Combined machine learning models demonstrated strong predictive performance, with Random Forest achieving the highest Area Under the Curve(AUC) scores for GC(0.94[0.83-1.00]), CRC (0.75[0.62-0.86]), and IBD (0.93[0.86-0.98]). These models were then employed for cross-disease analysis, revealing that models trained on GC data successfully predicted IBD biomarkers, while CRC models predicted GC biomarkers with optimal performance scores.
Conclusion: These findings emphasize the potential of microbial and metabolic profiling in cross-disease characterization particularly for GIDs, advancing biomarker discovery for improved diagnostics and targeted therapies.
Keywords: Biomarkers; Colorectal cancer; Gastric cancer; Inflammatory bowel disease; Machine learning; Metabolome; Microbiome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that this research was conducted without any competing personal or financial relationships that could be considered potential influences on the work presented in this paper.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

