3'UTR RNA editing driven by ADAR1 modulates MDM2 expression in breast cancer cells
- PMID: 40381037
- PMCID: PMC12085317
- DOI: 10.1007/s10142-025-01611-3
3'UTR RNA editing driven by ADAR1 modulates MDM2 expression in breast cancer cells
Abstract
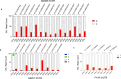
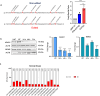
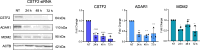
Epitranscriptomic changes in the transcripts of cancer related genes could modulate protein levels. RNA editing, particularly A-to-I(G) editing catalyzed by ADAR1, has been implicated in cancer progression. RNA editing events in the 3' untranslated region (3'UTR) can regulate mRNA stability, localization, and translation, underscoring the importance of exploring their impact in cancer. Here, we performed an in silico analysis to detect breast cancer enriched RNA editing sites using the TCGA breast cancer RNA-seq dataset. Notably, the majority of differential editing events mapped to 3' untranslated regions (3'UTRs). We confirmed A-to-I(G) editing in the 3'UTRs of MDM2 (Mouse Double Minute 2 homolog), GINS1 (GINS Complex Subunit 1), and F11R (Junctional Adhesion Molecule A) in breast cancer cells. RNA immunoprecipitation with ADAR1 antibody confirmed the interaction between ADAR1 and MDM2, GINS1, and F11R 3'UTRs. ADAR1 knockdown revealed decreased editing levels, establishing ADAR1 as the editing enzyme. A reporter assay for MDM2, an oncogene overexpressed mostly in luminal breast cancers, demonstrated that RNA editing enhances protein expression, in agreement with reduced MDM2 protein levels in ADAR1 knockdown cells. Further exploration into the mechanisms of 3'UTR editing events revealed an interaction between ADAR1 and CSTF2, a core component of the polyadenylation machinery, as identified through biotin-based proximity labeling mass spectroscopy, and co-immunoprecipitation experiments. Furthermore, CSTF2 knockdown reduced both ADAR1 and MDM2 protein levels. Our findings highlight implications for MDM2 regulation by ADAR1-dependent 3'UTR RNA editing and present an interplay between RNA editing on 3'UTRs and the mRNA polyadenylation machinery. These results improve our understanding of ADAR1's role in cancer-associated 3' UTR RNA editing and its potential as a therapeutic target.
Keywords: 3’UTR; ADAR1; CSTF2; F11R; GINS1; MDM2; Proximity biotinylation; RNA editing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
ADAR1-mediated RNA-editing of 3'UTRs in breast cancer.Biol Res. 2018 Oct 5;51(1):36. doi: 10.1186/s40659-018-0185-4. Biol Res. 2018. PMID: 30290838 Free PMC article.
-
ADAR1-mediated 3' UTR editing and expression control of antiapoptosis genes fine-tunes cellular apoptosis response.Cell Death Dis. 2017 May 25;8(5):e2833. doi: 10.1038/cddis.2017.12. Cell Death Dis. 2017. PMID: 28542129 Free PMC article.
-
Combinatory RNA-Sequencing Analyses Reveal a Dual Mode of Gene Regulation by ADAR1 in Gastric Cancer.Dig Dis Sci. 2018 Jul;63(7):1835-1850. doi: 10.1007/s10620-018-5081-9. Epub 2018 Apr 25. Dig Dis Sci. 2018. PMID: 29691780
-
ADAR1-mediated RNA editing in breast cancer: molecular mechanisms and therapeutic implications.Med Oncol. 2025 Aug 9;42(9):421. doi: 10.1007/s12032-025-02979-9. Med Oncol. 2025. PMID: 40782195 Review.
-
RNA editing and immune control: from mechanism to therapy.Curr Opin Genet Dev. 2024 Jun;86:102195. doi: 10.1016/j.gde.2024.102195. Epub 2024 Apr 20. Curr Opin Genet Dev. 2024. PMID: 38643591 Free PMC article. Review.
References
-
- Ayaz G, Yasar P, Olgun CE, Karakaya B, Kars G, Razizadeh N, Yavuz K, Turan G, Muyan M (2019) Dynamic transcriptional events mediated by estrogen receptor alpha. Front Biosci (Landmark Ed).;24(2):245–276. 10.2741/4716. PMID: 30468654 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

