Targeting MYOF suppresses pancreatic ductal adenocarcinoma progression by inhibiting ILF3-LCN2 signaling through disrupting OTUB1-mediated deubiquitination of ILF3
- PMID: 40381229
- PMCID: PMC12145718
- DOI: 10.1016/j.redox.2025.103665
Targeting MYOF suppresses pancreatic ductal adenocarcinoma progression by inhibiting ILF3-LCN2 signaling through disrupting OTUB1-mediated deubiquitination of ILF3
Abstract
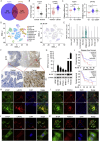
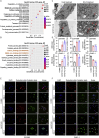
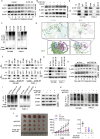
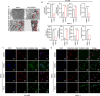
Pancreatic ductal adenocarcinoma (PDAC) is still a highly aggressive and fatal disease. The molecular mechanisms for PDAC progression are still not fully understood. Here, we demonstrated the overexpression of MYOF in PDAC in multiple sample sets, which is significantly associated with poor outcome of PDAC patients. MYOF knockout suppresses PDAC progression in vitro and in vivo. MYOF knockout exerts its effects by promoting ferroptosis via downregulating LCN2 expression. Ectopic LCN2 expression overcame the effects of MYOF knockout in PDAC cells. Mechanistically, MYOF respectively recruits OTUB1 and ILF3 to enhance their interaction and relieves ILF3 protein ubiquitination and degradtion. MYOF maintains ILF3 protein stability, thereby enhances ILF3 interacting with and improving LCN2 mRNA stability. Moreover, we screened and identified natural compound Picroside II potentially targets MYOF to suppress PDAC progression. These findings uncover the biological roles and mechanisms of MYOF and preliminarily indicate the potential of targeting MYOF in PDAC progression, highlighting a novel therapeutic strategy for PDAC.
Keywords: Ferroptosis; ILF3; LCN2; MYOF; OTUB1; Pancreatic cancer.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures












Similar articles
-
lncRNA THAP9-AS1 Promotes Pancreatic Ductal Adenocarcinoma Growth and Leads to a Poor Clinical Outcome via Sponging miR-484 and Interacting with YAP.Clin Cancer Res. 2020 Apr 1;26(7):1736-1748. doi: 10.1158/1078-0432.CCR-19-0674. Epub 2019 Dec 12. Clin Cancer Res. 2020. PMID: 31831555
-
Lipocalin-2 Promotes Pancreatic Ductal Adenocarcinoma by Regulating Inflammation in the Tumor Microenvironment.Cancer Res. 2017 May 15;77(10):2647-2660. doi: 10.1158/0008-5472.CAN-16-1986. Epub 2017 Mar 1. Cancer Res. 2017. PMID: 28249896 Free PMC article.
-
MGST1 facilitates novel KRASG12D inhibitor resistance in KRASG12D-mutated pancreatic ductal adenocarcinoma by inhibiting ferroptosis.Mol Med. 2024 Nov 5;30(1):199. doi: 10.1186/s10020-024-00972-y. Mol Med. 2024. PMID: 39501138 Free PMC article.
-
SIK1 promotes ferroptosis resistance in pancreatic cancer via HDAC5-STAT6-SLC7A11 axis.Cancer Lett. 2025 Jul 28;623:217726. doi: 10.1016/j.canlet.2025.217726. Epub 2025 Apr 16. Cancer Lett. 2025. PMID: 40250791
-
Glycolysis and tumor progression promoted by the m6A writer VIRMA via m6A-dependent upregulation of STRA6 in pancreatic ductal adenocarcinoma.Cancer Lett. 2024 May 28;590:216840. doi: 10.1016/j.canlet.2024.216840. Epub 2024 Apr 9. Cancer Lett. 2024. PMID: 38604311
References
-
- Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. - PubMed
-
- Cai J., et al. Advances in the epidemiology of pancreatic cancer: trends, risk factors, screening, and prognosis. Cancer Lett. 2021;520:1–11. - PubMed
-
- Midha S., Chawla S., Garg P.K. Modifiable and non-modifiable risk factors for pancreatic cancer: a review. Cancer Lett. 2016;381:269–277. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

