Targeting C1q prevents microglia-mediated synaptic removal in neuropathic pain
- PMID: 40382320
- PMCID: PMC12085617
- DOI: 10.1038/s41467-025-59849-1
Targeting C1q prevents microglia-mediated synaptic removal in neuropathic pain
Abstract
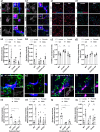
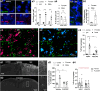
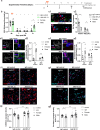
Activation of spinal microglia following peripheral nerve injury is a central component of neuropathic pain pathology. While the contributions of microglia-mediated immune and neurotrophic signalling have been well-characterized, the phagocytic and synaptic pruning roles of microglia in neuropathic pain remain less understood. Here, we show that peripheral nerve injury induces microglial engulfment of dorsal horn synapses, leading to a preferential loss of inhibitory synapses and a shift in the balance between inhibitory and excitatory synapse density. This synapse removal is dependent on the microglial complement-mediated synapse pruning pathway, as mice deficient in complement C3 and C4 do not exhibit synapse elimination. Furthermore, pharmacological inhibition of the complement protein C1q prevents dorsal horn inhibitory synapse loss and attenuates neuropathic pain. Therefore, these results demonstrate that the complement pathway promotes persistent pain hypersensitivity via microglia-mediated engulfment of dorsal horn synapses in the spinal cord, revealing C1q as a therapeutic target in neuropathic pain.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.Y., V.M., Y.A.Z., N.R.K.A., and T.Y. are employees of Annexon Inc., a venture-funded private biotechnology company. All other authors have no competing interests.
Figures



References
-
- Baron, R., Binder, A. & Wasner, G. Neuropathic pain: diagnosis, pathophysiological mechanisms, and treatment. Lancet Neurol.9, 807–819 (2010). - PubMed
-
- Inoue, K. & Tsuda, M. Microglia in neuropathic pain: cellular and molecular mechanisms and therapeutic potential. Nat. Rev. Neurosci.19, 138–152 (2018). - PubMed
-
- Gilmore, S. A. & Skinner, R. D. Intraspinal non-neuronal cellular responses to peripheral nerve injury. Anat. Rec.194, 369–387 (1979). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

