A global atlas and drivers of antimicrobial resistance in Salmonella during 1900-2023
- PMID: 40382325
- PMCID: PMC12085583
- DOI: 10.1038/s41467-025-59758-3
A global atlas and drivers of antimicrobial resistance in Salmonella during 1900-2023
Abstract
Although previous studies using phenotypic or/and genomic approaches monitoring have revealed the spatiotemporal distribution of antimicrobial resistance (AMR) in Salmonella in local areas, their geographical patterns and driving factors remain largely unknown at a global scale. Here, we performed an analysis of publicly available data of 208,233 Salmonella genomes in 148 countries/regions between 1900 and 2023 and explored driving indicators of AMR. Overall, we found that the geographic distribution of AMR varied depending on the location, source, and serovar. The proportion of AMR levels increased across six continents, especially in serovars Agona, Dublin, I 1,4,[5],12:i:-, Muenchen, Senftenberg, Mbandaka mainly from chickens, food, wild animals, and the environment, while decreased in Schwarzengrund and Saintpaul mainly from cattle, pigs, and turkeys. We also found that S. Typhimurium exhibiting macro, red, dry, and rough was detected as early as 1992 in the USA, earlier than in China. Moreover, we identified that antibiotic consumption, agriculture, climate, urban, health, and socioeconomic factors contribute to the development of AMR in Salmonella. We present a globally high-resolution genetic atlas of Salmonella and also identify some factors driving the rise of AMR, which can provide valuable information for understanding the transmission dynamics and evolutionary trajectories of Salmonella.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that there are no competing interests.
Figures



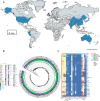
References
-
- World Health Organization. WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. https://www.who.int/publications/i/item/9789240093461/ (2024). - PubMed
-
- Ma, Y. et al. WHO revised bacterial priority pathogens list to encourage global actions to combat AMR. hLife2, 607–610 (2024).
-
- Xiao, Y. & Nishijima, T. Status and challenges of global antimicrobial resistance control: A dialogue between Professors Yonghong Xiao and Takeshi Nishijima. hLife2, 47–49 (2024).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

