Antiviral potential of diosmin against influenza A virus
- PMID: 40382364
- PMCID: PMC12085588
- DOI: 10.1038/s41598-025-00744-6
Antiviral potential of diosmin against influenza A virus
Abstract
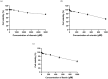
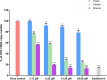
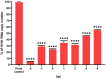
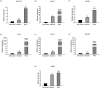
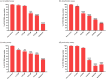
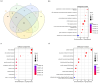
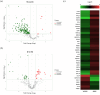
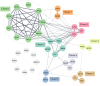
Influenza poses a global health threat. With drug-resistant strains emerging, there is an urgent need for effective antiviral drugs. This study explores antiviral potential of flavonoids against influenza A virus (IAV) and their mechanism of action. By utilizing in silico docking as a screening approach, diosmin, orientin, and fisetin were identified as flavonoids with the strongest interactions with viral proteins. Out of them, diosmin was found to effectively inhibit IAV replication in vitro, particularly at the attachment and post-entry stages, with significant inhibition observed at 0-h post-infection (hpi) and 2 hpi, while also demonstrated prophylactic activity, peaking at - 2 hpi. Following that, diosmin significantly increases the expression of antiviral genes, which may relate to the discovery of its prophylactic activity. Proteomics analysis showed that diosmin treatment during the post-entry stage of IAV replication reduced viral protein levels, confirming its antiviral activity at this point. Additionally, diosmin also modulated host proteins related to innate immunity, inducing type I interferon and anti-inflammatory responses during the infection. These findings provide preliminary evidence of diosmin's antiviral and prophylactic activity against IAV, paving the way for further research on its mechanism of action.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Unveiling the Antiviral Potential of Minocycline: Modulation of Nuclear Export of Viral Ribonuclear Proteins during Influenza Virus Infection.Viruses. 2024 Aug 18;16(8):1317. doi: 10.3390/v16081317. Viruses. 2024. PMID: 39205291 Free PMC article.
-
Novel Anti-Viral Properties of the Herbal Extract of Davallia mariesii against Influenza A Virus.Viruses. 2024 Mar 28;16(4):523. doi: 10.3390/v16040523. Viruses. 2024. PMID: 38675866 Free PMC article.
-
Hemagglutinin docking analysis-based visual detection of antiviral therapeutic efficacy of umifenovir against influenza virus.Biosens Bioelectron. 2025 Nov 1;287:117762. doi: 10.1016/j.bios.2025.117762. Epub 2025 Jul 7. Biosens Bioelectron. 2025. PMID: 40639140
-
Emerging cellular targets for influenza antiviral agents.Trends Pharmacol Sci. 2012 Feb;33(2):89-99. doi: 10.1016/j.tips.2011.10.004. Epub 2011 Dec 22. Trends Pharmacol Sci. 2012. PMID: 22196854 Review.
-
Targeting the Ubiquitin Proteasome System to Combat Influenza A Virus: Hijacking the Cleanup Crew.Rev Med Virol. 2024 Nov;34(6):e70005. doi: 10.1002/rmv.70005. Rev Med Virol. 2024. PMID: 39516190 Review.
References
-
- Vaidya, B. et al. Effectiveness of periodic treatment of quercetin against influenza A virus H1N1 through modulation of protein expression. J. Agric. Food Chem.64, 4416–4425. 10.1021/acs.jafc.6b00148 (2016). - PubMed
-
- Eccleston-Turner, M., Phelan, A. & Katz, R. Preparing for the next pandemic—The WHO’s global influenza strategy. N. Engl. J. Med.381, 2192–2194. 10.1056/NEJMp1905224 (2019). - PubMed
-
- James, S. H. & Whitley, R. J. in Infectious Diseases. 4th edn (eds Jonathan Cohen, William G. Powderly, & Steven M. Opal) 1465–1471.e1461 (Elsevier, 2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

