Microsporidian infection of mosquito larvae changes the host-associated microbiome towards the synthesis of antimicrobial factors
- PMID: 40382661
- PMCID: PMC12085064
- DOI: 10.1186/s13071-025-06813-z
Microsporidian infection of mosquito larvae changes the host-associated microbiome towards the synthesis of antimicrobial factors
Abstract
Background: Microsporidians (Microsporidia) are a group of obligate intracellular parasites that commonly infect mosquitoes. Recently, it has been shown that infection by these parasites can alter the composition and functionality of the mosquito-associated microbiome. The host-associated microbiome of the mosquito can play a pivotal role in various physiological processes of this host, including its vector competence for pathogens. Thus, understanding how microsporidians shape the mosquito microbiome may be crucial for elucidating interactions between these parasites and their mosquito hosts, which are also vectors for other parasites and pathogens.
Methods: The effects of microsporidian infection on the microbiome structure and functionality of Culex pipiens and Culex torrentium larvae under semi-natural conditions were examined. The host-associated microbiome of Cx. pipiens (n = 498) and Cx. torrentium (n = 465) larvae, including that of the 97 infected individuals of these samples, was analysed using 16S DNA profiling and functional prediction analysis.
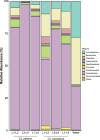
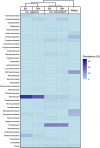
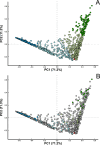
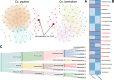
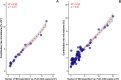
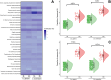
Results: Microbiome network analysis revealed that, in the microsporidian-positive larvae, host microbial communities consistently grouped within a common bacterial module that included Aerococcaceae, Lactobacillaceae, Microbacteriaceae, Myxococcaceae, and Polyangiaceae. Indicator species analysis revealed two strong positive correlations between microsporidian infection and the presence of Weissella cf. viridescens and Wolbachia pipientis. Functional predictions of microbiome content showed enrichment in biosynthetic pathways for ansamycin and vancomycin antibiotic groups in infected larvae. Furthermore, the MexJK-OprM multidrug-resistance module was exclusively present in the infected larvae, while carbapenem- and vancomycin-resistance modules were specific to the microsporidian-free larvae.
Conclusions: Our results demonstrate that microsporidian infection alters the microbial community composition in mosquito larvae. Moreover, they show that microsporidian infection can increase the antimicrobial capabilities of the host-associated microbiome. These results provide novel insights into host microbiome-parasite interactions and have potential implications for the vector competencies of mosquitoes.
Keywords: Weissella viridescens; Wolbachia pipientis; Antimicrobial factors; Biosynthetic pathways; Disease vectors; Host–microbe interactions; Microbiota; Microsporidia; Mosquitoes; Parasites.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures
Similar articles
-
Microsporidian Infection in Mosquitoes (Culicidae) Is Associated with Gut Microbiome Composition and Predicted Gut Microbiome Functional Content.Microb Ecol. 2023 Jan;85(1):247-263. doi: 10.1007/s00248-021-01944-z. Epub 2021 Dec 23. Microb Ecol. 2023. PMID: 34939130 Free PMC article.
-
Nested patterns of commensals and endosymbionts in microbial communities of mosquito vectors.BMC Microbiol. 2024 Oct 26;24(1):434. doi: 10.1186/s12866-024-03593-x. BMC Microbiol. 2024. PMID: 39455905 Free PMC article.
-
Temporal Variations of Microbiota Associated with the Immature Stages of Two Florida Culex Mosquito Vectors.Microb Ecol. 2017 Nov;74(4):979-989. doi: 10.1007/s00248-017-0988-9. Epub 2017 May 11. Microb Ecol. 2017. PMID: 28492989
-
Wolbachia dominance influences the Culex quinquefasciatus microbiota.Sci Rep. 2023 Nov 3;13(1):18980. doi: 10.1038/s41598-023-46067-2. Sci Rep. 2023. PMID: 37923779 Free PMC article. Review.
-
Curious entanglements: interactions between mosquitoes, their microbiota, and arboviruses.Curr Opin Virol. 2019 Aug;37:26-36. doi: 10.1016/j.coviro.2019.05.005. Epub 2019 Jun 5. Curr Opin Virol. 2019. PMID: 31176069 Free PMC article. Review.
References
-
- Joos R, Boucher K, Lavelle A, Arumugam M, Blaser MJ, Claesson MJ, et al. Examining the healthy human microbiome concept. Nat Rev Microbiol. 2024;23:192–205. 10.1038/s41579-024-01107-0. - PubMed
-
- Lee J-Y, Tsolis RM, Bäumler AJ. The microbiome and gut homeostasis. Science. 2022;377(6601):eabp9960. 10.1126/science.abp9960. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

