LncRNA-induced lysosomal localization of NHE1 promotes increased lysosomal pH in macrophages leading to atherosclerosis
- PMID: 40383150
- PMCID: PMC12192704
- DOI: 10.1016/j.jbc.2025.110246
LncRNA-induced lysosomal localization of NHE1 promotes increased lysosomal pH in macrophages leading to atherosclerosis
Abstract
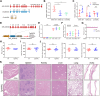
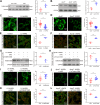
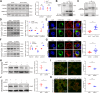
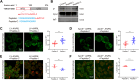
ANRIL, also referred to as CDKN2B-AS1, is an lncRNA gene implicated in the pathogenesis of multiple human diseases including atherosclerotic coronary artery disease; however, definitive in vivo evidence is lacking and the underlying molecular mechanism is largely unknown. In this study, we show that ANRIL overexpression causes atherosclerosis in vivo as transgenic mouse overexpression of full-length ANRIL (NR_003529) increases inflammation and aggravates atherosclerosis under ApoE-/- background (ApoE-/-ANRIL mice). Mechanistically, ANRIL reduces the expression of miR-181b-5p, which leads to increased TMEM106B expression. TMEM106B is significantly upregulated in the atherosclerotic lesions of both human CAD patients and ApoE-/-ANRIL mice. TMEM106B interacts and colocalizes with Na+-H+ exchanger NHE1, which results in the mislocalization of NHE1 from cell membranes to lysosomal membranes, leading to increased lysosomal pH in macrophages. Large truncation and point mutation analyses define the critical amino acids for TMEM106B-NHE1 interaction and lysosomal pH regulation as F115 and F117 on TMEM106B and I537, C538, and G539 on NHE1. Topological analysis suggests that both N terminus and C terminus of NHE1 are located inside lysosomal lumen, consistent with our finding that NHE1 is an important new proton efflux channel involved in raising lysosomal pH. A short TMEM106B peptide (YGRKKRRQRRR-L111A112V113F114F115L116F117) disrupting the TMEM106B-NHE1 interaction normalized lysosomal pH in macrophages with ANRIL overexpression. Our data demonstrate that ANRIL promotes atherosclerosis in vivo and identify the ANRIL-miR-181b-5p-TMEM106B-NHE1-lysosomal pH axis as the underlying molecular pathogenic mechanism for the chromosome 9p21.3 genetic locus for coronary artery disease.
Keywords: ANRIL; NHE1; TMEM106B; atherosclerosis; coronary artery disease; lysosomal pH; macrophage; miR-181b-5p.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





Similar articles
-
SGLT2i Alleviates Atherosclerosis by Inhibiting NHE1 Activation to Protect against Macrophage Senescence Induced by Angiotensin II.Comb Chem High Throughput Screen. 2025;28(10):1754-1765. doi: 10.2174/0113862073310500240514045321. Comb Chem High Throughput Screen. 2025. PMID: 38803180
-
The 9p21.3 Coronary Artery Disease Risk Locus Drives Vascular Smooth Muscle Cells to an Osteochondrogenic State.Arterioscler Thromb Vasc Biol. 2025 May;45(5):702-721. doi: 10.1161/ATVBAHA.124.322045. Epub 2025 Mar 27. Arterioscler Thromb Vasc Biol. 2025. PMID: 40143812 Free PMC article.
-
Galactin-8 DNA methylation mediates macrophage autophagy through the MAPK/mTOR pathway to alleviate atherosclerosis.Sci Rep. 2025 Jan 2;15(1):603. doi: 10.1038/s41598-024-85036-1. Sci Rep. 2025. PMID: 39747459 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Role of long non-coding RNAs in neurofibromatosis and Schwannomatosis: pathogenesis and therapeutic potential.Epigenomics. 2024 Dec-Dec;16(23-24):1453-1464. doi: 10.1080/17501911.2024.2430170. Epub 2024 Nov 27. Epigenomics. 2024. PMID: 39601046 Review.
References
-
- Weber C., Habenicht A.J.R., von Hundelshausen P. Novel mechanisms and therapeutic targets in atherosclerosis: inflammation and beyond. Eur. Heart J. 2023;44:2672–2681. - PubMed
-
- Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

