Similarity of immune-associated markers in COVID-19 and Kawasaki disease: analyses from bioinformatics and machine learning
- PMID: 40383755
- PMCID: PMC12087065
- DOI: 10.1186/s12887-025-05752-z
Similarity of immune-associated markers in COVID-19 and Kawasaki disease: analyses from bioinformatics and machine learning
Abstract
Background: Infection by the SARS-CoV-2 virus can cause coronavirus disease 2019 (COVID-19) and can also exacerbate the symptoms of Kawasaki disease (KD), an acute vasculitis that mostly affects children. This study used bioinformatics and machine learning to examine similarities in the molecular pathogenesis of COVID-19 and KD.
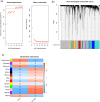
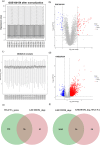
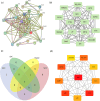
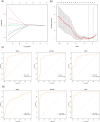
Methods: We first identified disease-associated modules in KD using weighted gene co-expression network analysis. Then, we determined shared differentially expressed genes (DEGs) in training datasets for KD (GSE100154) and COVID-19 (GSE225220), performed functional annotation of these shared DEGs, and used Cytoscape plug-ins (MCODE and Cytohubba) to characterize the protein-protein interaction (PPI) network and identify the hub genes. We performed Least Absolute Shrinkage and Selection Operator(LASSO) regression and receiver operating characteristic (ROC) curve analysis to identify the most robust markers, validated these results by analysis of two other datasets (GSE73461 and GSE18606), and then calculated the correlations of these key genes with immune cells.
Results: This analysis identified 26 shared DEGs in COVID-19 and KD. The results from functional annotation showed that the shared DEGs primarily functioned in immune responses, the formation of neutrophil extracellular traps, and NOD-like receptor signaling pathways. There were three key genes (PGLYRP1, DEFA4, RETN), and they had positive correlations with monocytes, M0 macrophages, and dendritic cells, which function as immune infiltrating cells in KD.
Conclusion: The potential immune-associated biomarkers (PGLYRP1, DEFA4, RETN) along with their shared pathways, hold promise for advancing investigations into the underlying pathogenesis of KD and COVID-19.
Keywords: Bioinformatics analysis; COVID-19; Immune cell infiltration; Kawasaki disease (KD); Weighted gene co-expression network analysis (WGCNA).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Identification of hub biomarkers and immune-related pathways participating in the progression of Kawasaki disease by integrated bioinformatics analysis.Immunobiology. 2023 Nov;228(6):152750. doi: 10.1016/j.imbio.2023.152750. Epub 2023 Sep 26. Immunobiology. 2023. PMID: 37837870
-
Bioinformatic analysis of underlying mechanisms of Kawasaki disease via Weighted Gene Correlation Network Analysis (WGCNA) and the Least Absolute Shrinkage and Selection Operator method (LASSO) regression model.BMC Pediatr. 2023 Feb 24;23(1):90. doi: 10.1186/s12887-023-03896-4. BMC Pediatr. 2023. PMID: 36829193 Free PMC article.
-
Identification of Hub Biomarkers and Immune and Inflammation Pathways Contributing to Kawasaki Disease Progression with RT-qPCR Verification.J Immunol Res. 2023 Apr 6;2023:1774260. doi: 10.1155/2023/1774260. eCollection 2023. J Immunol Res. 2023. PMID: 39670237 Free PMC article.
-
Identification of Key Genes and Underlying Mechanisms in Acute Kawasaki Disease Based on Bioinformatics Analysis.Med Sci Monit. 2021 Jul 22;27:e930547. doi: 10.12659/MSM.930547. Med Sci Monit. 2021. PMID: 34290221 Free PMC article.
-
Phenotype, Susceptibility, Autoimmunity, and Immunotherapy Between Kawasaki Disease and Coronavirus Disease-19 Associated Multisystem Inflammatory Syndrome in Children.Front Immunol. 2021 Feb 26;12:632890. doi: 10.3389/fimmu.2021.632890. eCollection 2021. Front Immunol. 2021. PMID: 33732254 Free PMC article.
References
-
- Newburger JW, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a statement for health professionals from the committee on rheumatic fever, endocarditis and Kawasaki disease, Council on cardiovascular disease in the young. Am Heart Association Circulation. 2004;110(17):2747–71. 10.1161/01.cir.0000145143.19711.78. - PubMed
-
- McCrindle BW, et al. Diagnosis, treatment, and Long-Term management of Kawasaki disease: A scientific statement for health professionals from the American heart association. Circulation. 2017;135(17):e927–99. 10.1161/cir.0000000000000484. - PubMed
-
- Harnden A, Tulloh R, Burgner D. Kawasaki disease. BMJ. 2014;349:g5336. 10.1136/bmj.g5336. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

