A broad antibody with enhanced HIV-1 neutralization via bispecific antibody-mediated prepositioning
- PMID: 40383778
- PMCID: PMC12086220
- DOI: 10.1038/s41467-025-60035-6
A broad antibody with enhanced HIV-1 neutralization via bispecific antibody-mediated prepositioning
Abstract
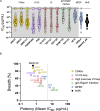
Antibodies targeting the highly conserved prehairpin intermediate (PHI) of class I viral membrane-fusion proteins are generally weakly neutralizing and are not considered viable therapeutic agents. We previously demonstrated that antibodies targeting the gp41 N-heptad repeat (NHR), which is transiently exposed in the HIV-1 PHI, exhibit enhanced broad neutralization in cells expressing the Fc receptor, FcγRI. To enhance neutralization in cells lacking FcγRI, we here develop a bispecific antibody (bsAb) by fusing an NHR-targeting antibody to an antibody against CD4, the HIV-1 receptor on T cells. The bsAb provides a 5000-fold neutralization enhancement and shows unprecedented neutralization breadth compared to existing broadly neutralizing antibodies. Importantly, the bsAb reduces viral load in HIV-1-infected humanized male mice, and viral envelope sequencing under bsAb pressure revealed an NHR mutation that potentially impairs viral fitness. These findings validate the NHR as a potential HIV-1 therapeutic target, setting the stage for a new class of broadly neutralizing antibodies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Orloff, G. M., Orloff, S. L., Kennedy, M. S., Maddon, P. J. & McDougal, J. S. Penetration of CD4 T cells by HIV-1. The CD4 receptor does not internalize with HIV, and CD4-related signal transduction events are not required for entry. J. Immunol.146, 2578–2587 (1991). - PubMed
-
- Moore, J. P., Trkola, A. & Dragic, T. Co-receptors for HIV-1 entry. Curr. Opin. Immunol.9, 551–562 (1997). - PubMed
-
- Chan, D. C. & Kim, P. S. HIV entry and its inhibition. Cell93, 681–684 (1998). - PubMed
-
- Eckert, D. M. & Kim, P. S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem.70, 777–810 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

