Transcriptome sequencing of Antheraea pernyi antennae for identification of olfactory-related genes
- PMID: 40383784
- PMCID: PMC12087209
- DOI: 10.1186/s12864-025-11698-4
Transcriptome sequencing of Antheraea pernyi antennae for identification of olfactory-related genes
Abstract
Background: In insects, the olfactory system governs physiological and behavioral processes by detecting various odorous molecules. Despite its economic importance and adaptability, the olfactory mechanism of Antheraea pernyi remains insufficiently understood, limiting its potential for pest management and as a model organism. Hence, we aimed to conduct transcriptome sequencing to explore olfactory-related genes in the antennae, serving as the most important olfactory organ in adult A. pernyi.
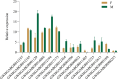
Results: Based on the datasets, 1184 differently expressed genes (DEGs), including 484 upregulated and 700 downregulated genes, were identified by comparing the transcriptome profiles of the male and female antennae of A. pernyi. Moreover, 20, 7, 30, 11, and 2 candidate genes encoding odorant-binding proteins (OBPs), chemosensory proteins (CSPs), odorant receptors (ORs), ionotropic receptors (IRs), and sensory neuron membrane proteins (SNMPs), respectively, involved in pheromone perception, odor binding, pesticide resistance, and growth and development regulation were screened, and most of which were expressed in both male and female antennae while the expression levels of these candidate genes varied significantly between males and females. Multiple sequence alignment indicated that the six OBPs exhibited typical characteristics, containing six conserved Cys residues with the sequence of C1-X26-30-C2-X3-C3-X41-42-C4-X8-10-C5-X8-C6. All CSPs followed a highly conserved pattern with four Cys residues arranged with an exact spacing of C1-X6-C2-X18-19-C3-X2-C4. Different numbers of transmembrane domains were found in ORs, IRs, and SNMPs. In addition, several DEGs involve signal transduction underlying chemoreception were also identified from the transcriptome data, including guanine nucleotide-binding protein (G protein), cGMP-dependent protein kinase (PKA), calmodulin-A (CaM-A), mitogen-activated protein kinase 1 (MAPK1), and phospholipase D2 (PLD2).
Conclusion: This study enriches the olfactory gene database of A. pernyi, providing insights into olfactory mechanisms crucial for mating and pest control, with implications for enhancing breeding strategies and ensuring the sustainability of the silk industry. These findings may serve as a theoretical foundation for a better understanding of the olfactory mechanisms of A. pernyi.
Keywords: Antheraea pernyi; Antennae; Bioinformatics analysis; Olfactory-related genes; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae).Insect Biochem Mol Biol. 2014 Sep;52:69-81. doi: 10.1016/j.ibmb.2014.06.006. Epub 2014 Jul 3. Insect Biochem Mol Biol. 2014. PMID: 24998398
-
Identification and characterization of chemosensory genes in the antennal transcriptome of Spodoptera exigua.Comp Biochem Physiol Part D Genomics Proteomics. 2018 Sep;27:54-65. doi: 10.1016/j.cbd.2018.05.001. Epub 2018 May 7. Comp Biochem Physiol Part D Genomics Proteomics. 2018. PMID: 29787920
-
Antennal transcriptome analyses and olfactory protein identification in an important wood-boring moth pest, Streltzoviella insularis (Lepidoptera: Cossidae).Sci Rep. 2019 Nov 29;9(1):17951. doi: 10.1038/s41598-019-54455-w. Sci Rep. 2019. PMID: 31784624 Free PMC article.
-
Identification of chemosensory genes from the antennal transcriptome of Indian meal moth Plodia interpunctella.PLoS One. 2018 Jan 5;13(1):e0189889. doi: 10.1371/journal.pone.0189889. eCollection 2018. PLoS One. 2018. PMID: 29304134 Free PMC article.
-
Sex pheromones and olfactory proteins in Antheraea moths: A. pernyi and A. polyphemus (Lepidoptera: Saturniidae).Arch Insect Biochem Physiol. 2020 Oct;105(2):e21729. doi: 10.1002/arch.21729. Epub 2020 Aug 6. Arch Insect Biochem Physiol. 2020. PMID: 32761939 Review.
References
-
- Yang DQ, Li DL, Jiang LL, Lin J, Yue GQ, Xiao K, Hao XX, Ji Q, Hong YC, Cai PM, Yang JQ. Antennal transcriptome analysis of Psyttalia incisi (silvestri) (Hymenoptera: Braconidae): identification and tissue expression profiling of candidate odorant-binding protein genes. Mol Biol Rep. 2024;51(1):333. - PubMed
-
- Brito NF, Moreira MF, Melo ACA. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51–65. - PubMed
-
- Kuang YH, Shangguan CZ, Yuan SC, Zhang QQ, Qiu ZY, Gao LW, Yu XD. Candidate odorant-binding protein and chemosensory protein genes in the turnip aphid Lipaphis erysimi. Arch Insect Biochem Physiol. 2023;113(4): e22022. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

