An energy metabolism-related signature relevant to the tumor immune microenvironment in HNSCC
- PMID: 40383838
- PMCID: PMC12086135
- DOI: 10.1007/s12672-025-02652-7
An energy metabolism-related signature relevant to the tumor immune microenvironment in HNSCC
Abstract
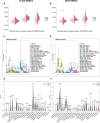
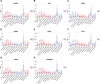
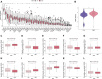
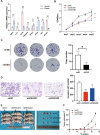
The importance of energy metabolism in cancer was explored by accumulating studies. Energy metabolism can affect the cellular activities of tumors. However, there is few research exploring the role of energy metabolism in tumor immune microenvironment. In this context, we constructed a novel energy metabolism-related prognostic signature containing 8 genes. The risk score calculated by the signature was analyzed to be an independent value of head and neck squamous cell carcinoma (HNSCC). We further validated the effectiveness and accuracy of our signature in The Cancer Genome Atlas Program (TCGA) cohort and Gene Expression Omnibus (GEO) cohort. Moreover, we also revealed a negative correlation between the risk score and the activity of the immune processes. Finally, we validated the function of Desmoglein 2 protein (DSG2), a risk gene in the signature, in tumor progression and found that knockdown of DSG2 remarkably suppressed the proliferation and migration of HNSCC cells, which further validated our analysis. In conclusion, the energy metabolism-related gene signature we built is a prospective biomarker of HNSCC, which can offer valuable clues for the research and development of immunotherapeutic drugs in HNSCC.
Keywords: DSG2; Energy metabolism; Head and neck squamous cell carcinoma; Prognostic signature; Tumor immune microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: All procedures in our study involving human tissues and animals adhered to the ethical standards set by the Institutional Ethics Committee of The Fourth Hospital of Changsha, as well as the 1964 Helsinki Declaration and its subsequent amendments. The code for the animal study protocol was CSSDSYY-YXLL-SC-2023-03-03, while the code for the human study protocol was CSSDSYY-YXLL-SC-2023-03-163. Written informed consent was obtained from all patients. Conflicts of interest: The authors declare no competing interests. Competing interests: The authors declare no competing interests.
Figures








Similar articles
-
Development and Validation of a Cholesterol-related Gene Signature for Prognostic Assessment in Head and Neck Squamous Cell Carcinoma.Curr Med Chem. 2024 Oct 15. doi: 10.2174/0109298673336147241010065340. Online ahead of print. Curr Med Chem. 2024. PMID: 39411955
-
Identification and verification of eight cancer-associated fibroblasts related genes as a prognostic signature for head and neck squamous cell carcinoma.Heliyon. 2023 Feb 28;9(3):e14003. doi: 10.1016/j.heliyon.2023.e14003. eCollection 2023 Mar. Heliyon. 2023. PMID: 36938461 Free PMC article.
-
Identification and validation of a prognostic signature of autophagy, apoptosis and pyroptosis-related genes for head and neck squamous cell carcinoma: to imply therapeutic choices of HPV negative patients.Front Immunol. 2023 Jan 10;13:1100417. doi: 10.3389/fimmu.2022.1100417. eCollection 2022. Front Immunol. 2023. PMID: 36703967 Free PMC article.
-
Bioinformatic analysis and verification of a lipid metabolism-related long noncoding RNA prognostic signature for head and neck squamous cell carcinoma.Cell Signal. 2023 Dec;112:110903. doi: 10.1016/j.cellsig.2023.110903. Epub 2023 Oct 7. Cell Signal. 2023. PMID: 37813294
-
Identification of pyroptosis-related gene prognostic signature in head and neck squamous cell carcinoma.Cancer Med. 2022 Dec;11(24):5129-5144. doi: 10.1002/cam4.4825. Epub 2022 May 16. Cancer Med. 2022. PMID: 35574984 Free PMC article.
References
-
- Weiss J, Hayes DN. Classifying squamous cell carcinoma of the head and neck: prognosis, prediction and implications for therapy. Expert Rev Anticancer Ther. 2014;14(2):229–36. - PubMed
-
- Van den Bossche V, et al. Microenvironment-driven intratumoral heterogeneity in head and neck cancers: clinical challenges and opportunities for precision medicine. Drug Resist Updates. 2022;60: 100806. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
