TRIM16 mediates secretory autophagy in head and neck cancer-associated fibroblasts
- PMID: 40383937
- PMCID: PMC12362711
- DOI: 10.1080/15548627.2025.2508064
TRIM16 mediates secretory autophagy in head and neck cancer-associated fibroblasts
Abstract
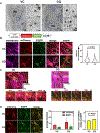
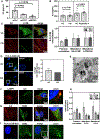
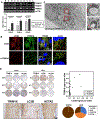
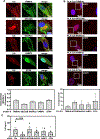
Improving treatment options for head and neck squamous cell carcinoma (HNSCC) requires a deeper understanding of the tumor microenvironment, particularly cancer-associated fibroblasts (CAFs). We previously reported that HNSCC-derived FGF2/bFGF (fibroblast growth factor 2) triggers cytokine release from CAFs via secretory autophagy. Here, using transmission electron microscopy, live-cell imaging, and immunofluorescence, we show that CAF autophagosomes transport cargo, including IL6, to the plasma membrane for secretion. Autophagy in CAFs is constitutive and independent of STAT3, MAPK1/ERK2-MAPK3/ERK1 and phosphoinositide 3-kinase (PI3K) signaling. Despite the significant role of secretory autophagy in CAFs, its molecular machinery has remained elusive. Using both a literature based, and an unbiased approach, we studied the molecular machinery involved in autophagosome trafficking in CAFs. We identified TRIM16, a protein previously reported to traffic to autophagosomes, upregulated in CAFs compared to normal oral fibroblasts. Immunohistochemistry of patient HNSCC stroma revealed co-expression of TRIM16 and LC3B, linking TRIM16 to autophagosome function. An unbiased proteomics profiling of immunoprecipitated LC3B+ vesicles in primary HNSCC CAFs revealed enrichment in trafficking proteins, focal adhesion, and mitochondrial proteins. We demonstrate that SEC22B, SNAP23, VAMP3, and STX4 colocalize with LC3B, IL6, and TRIM16 in CAFs. TRIM16 knockdown reduced autophagosomes at the plasma membrane and decreased IL6 secretion from CAFs. These findings uncover key molecular components involved in autophagy-mediated IL6 secretion in CAFs and suggest potential therapeutic targets for HNSCC.Abbreviations: ACTA2/αSMA: actin alpha 2, smooth muscle; CAF: cancer-associated fibroblasts; CM: conditioned media; CQ: chloroquine; DAPI: 4',6-diamidino-2-phenylindole; DMSO: dimethylsulfoxide; EGFP: enhanced green fluorescent protein; ELISA: enzyme-linked immunosorbent assay; ER: endoplasmic reticulum; FGF2/bFGF: fibroblast growth factor 2; FGFR: fibroblast growth factor receptor; GO: gene ontology; GORASP2/GRASP55: golgi reassembly stacking protein 2; HMGB1: high mobility group box 1; HNSCC: head and neck squamous cell carcinoma; HPV: human papillomavirus; IL6: interleukin 6; IP: immunoprecipitation; LC-MS/MS: liquid chromatography-mass spectrometry/mass spectrometry; LIR: LC3-interacting region; MAP1LC3B/LC3B: microtubule associated protein 1 light chain 3 beta; MAPK1/ERK2: mitogen-activated protein kinase 1; MAPK3/ERK1: mitogen-activated protein kinase 3; NFs: normal oral fibroblasts; NSCLC: non-small cell lung cancer; PLA: proximity ligation assay; SQSTM1/p62: sequestosome 1; STAT3: signal transducer and activator of transcription 3; SNAP23: synaptosome associated protein 23; SNARE: soluble N-ethyl-maleimide-sensitive factor attachment protein receptor; STX4: syntaxin 4; TEM: transmission electron microscopy; TGFB1: transforming growth factor beta 1; TMA: tissue microarray; TRIM: tri-partite motif; VAMP: vesicle associated membrane protein; VC: vehicle control.
Keywords: Autophagosome protein profiling; CAF; HNSCC; IL6; secretion; vesicular trafficking.
Conflict of interest statement
Disclosure statement
No potential conflict of interest was reported by the author(s).
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
