Deuteration of proteins boosted by cell lysates: high-resolution amide and H α magic-angle-spinning (MAS) NMR without the reprotonation bottleneck
- PMID: 40384771
- PMCID: PMC12082565
- DOI: 10.5194/mr-5-33-2024
Deuteration of proteins boosted by cell lysates: high-resolution amide and H α magic-angle-spinning (MAS) NMR without the reprotonation bottleneck
Abstract
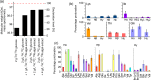
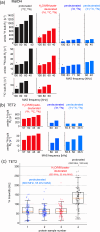
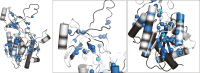
Amide-proton-detected magic-angle-spinning NMR of deuterated proteins has become a main technique in NMR-based structural biology. In standard deuteration protocols that rely on D O-based culture media, non-exchangeable amide sites remain deuterated, making these sites unobservable. Here we demonstrate that proteins produced with a H O-based culture medium doped with deuterated cell lysate allow scientists to overcome this "reprotonation bottleneck" while retaining a high level of deuteration (ca. 80 %) and narrow linewidths. We quantified coherence lifetimes of several proteins prepared with this labeling pattern over a range of magic-angle-spinning (MAS) frequencies (40-100 kHz). We demonstrate that under commonly used conditions (50-60 kHz MAS), the amide H linewidths with our labeling approach are comparable to those of perdeuterated proteins and better than those of protonated samples at 100 kHz. For three proteins in the 33-50 kDa size range, many previously unobserved amides become visible. We report how to prepare the deuterated cell lysate for our approach from fractions of perdeuterated cultures which are usually discarded, and we show that such media can be used identically to commercial media. The residual protonation of H sites allows for well-resolved H -detected spectra and H resonance assignment, exemplified by the de novo assignment of 168 H sites in a 39 kDa protein. The approach based on this H O/cell-lysate deuteration and MAS frequencies compatible with 1.3 or 1.9 mm rotors presents a strong sensitivity benefit over 0.7 mm 100 kHz MAS experiments.
Copyright: © 2024 Federico Napoli et al.
Conflict of interest statement
At least one of the (co-)authors is a member of the editorial board of Magnetic Resonance. The peer-review process was guided by an independent editor, and the authors also have no other competing interests to declare.
Figures



Similar articles
-
Sensitivity and resolution of proton detected spectra of a deuterated protein at 40 and 60 kHz magic-angle-spinning.J Biomol NMR. 2015 Feb;61(2):161-71. doi: 10.1007/s10858-015-9904-0. Epub 2015 Feb 8. J Biomol NMR. 2015. PMID: 25663049
-
100 kHz MAS Proton-Detected NMR Spectroscopy of Hepatitis B Virus Capsids.Front Mol Biosci. 2019 Jul 24;6:58. doi: 10.3389/fmolb.2019.00058. eCollection 2019. Front Mol Biosci. 2019. PMID: 31396521 Free PMC article.
-
Deuteration for High-Resolution Detection of Protons in Protein Magic Angle Spinning (MAS) Solid-State NMR.Chem Rev. 2022 May 25;122(10):10019-10035. doi: 10.1021/acs.chemrev.1c00681. Epub 2021 Dec 6. Chem Rev. 2022. PMID: 34870415 Review.
-
Is protein deuteration beneficial for proton detected solid-state NMR at and above 100 kHz magic-angle spinning?Solid State Nucl Magn Reson. 2017 Oct;87:126-136. doi: 10.1016/j.ssnmr.2017.07.004. Epub 2017 Jul 25. Solid State Nucl Magn Reson. 2017. PMID: 28802890
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
Cited by
-
Bumps on the Road: The Way to Clean Relaxation Dispersion Magic-Angle Spinning NMR.J Am Chem Soc. 2025 Aug 13;147(32):29315-29326. doi: 10.1021/jacs.5c09057. Epub 2025 Aug 1. J Am Chem Soc. 2025. PMID: 40748291 Free PMC article.
References
-
- Agarwal V, Penzel S, Szekely K, Cadalbert R, Testori E, Oss A, Past J, Samoson A, Ernst M, Böckmann A, Meier B H. De novo 3D structure determination from sub-milligram protein samples by solid-state 100 kHz MAS NMR spectroscopy. Angew Chem Int Edit. 2014;53:12253–12256. - PubMed
-
- Andreas LB, Stanek J, Le Marchand T, Bertarello A, Paepe DC-D, Lalli D, Krejčíková M, Doyen C, Öster C, Knott B, Wegner S, Engelke F, Felli IC, Pierattelli R, Dixon NE, Emsley L, Herrmann T, Pintacuda G. Protein residue linking in a single spectrum for magic-angle spinning NMR assignment. J Biomol NMR. 2015;62:253–261. - PubMed
-
- Andreas LB, Jaudzems K, Stanek J, Lalli D, Bertarello A, Le Marchand T, Cala-De Paepe D, Kotelovica S, Akopjana I, Knott B, Wegner S, Engelke F, Lesage A, Emsley L, Tars K, Herrmann T, Pintacuda G. Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. P Natl Acad Sci USA. 2016;113:9187–9192. - PMC - PubMed
LinkOut - more resources
Full Text Sources
