A metaproteomic analysis of the piglet fecal microbiome across the weaning transition
- PMID: 40384785
- PMCID: PMC12082470
- DOI: 10.3389/fmicb.2025.1504433
A metaproteomic analysis of the piglet fecal microbiome across the weaning transition
Abstract
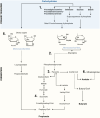
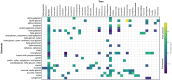
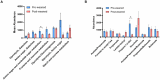
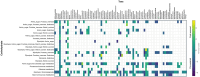
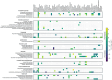
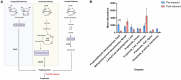
Microbiome analysis has relied largely on metagenomics to characterize microbial populations and predict their functions. Here, we used a metaproteomic analysis of the fecal microbiome in piglets before and after weaning to compare protein abundances as they pertain to microbial populations specific to either a milk- or plant-based diet. Fecal samples were collected from six piglets on the day of weaning and 4 weeks after transitioning to a standard nursery diet. Using the 12,554 protein groups identified in samples, we confirmed the shift in protein composition that takes place in response to the microbial succession following weaning and demonstrated the redundancy in metabolic processes between taxa. We identified taxa with roles as primary degraders based on corresponding proteins synthesized, thereby providing evidence for cross-feeding. Proteins associated with the breakdown of milk-specific carbohydrates were common among pre-weaned pigs, whereas the proteome of post-weaned piglets contained a greater abundance of proteins involved in the breaking down plant-specific carbohydrates. Furthermore, output revealed that production of propionate takes place via the propionaldehyde pathway in pre-weaned piglets, but changes to production via the succinate pathway in post-weaned piglets. Finally, a disproportionate quantity of carbohydrate-active enzymes (CAZymes) (~8%) were produced by fungi, which typically only represent ~0.1% of the microbiome taxa. Information gathered through this characterization of the metaproteome before and after weaning revealed important differences regarding the role of members in the microbial community, thereby providing information for the optimization of diets and products for both piglet and microbiome health.
Keywords: digestion; metaproteome; microbiome; swine; weaning.
Copyright © 2025 Rivera, Harlow, Cole, O’Meally, Garrett, Xiong, Oliver, Wells, Summers, Chhetri, Postnikova, Rempel, Crouse, Neville and Davies.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

