Minimal Residual Disease Detection: Implications for Clinical Diagnosis and Cancer Patient Treatment
- PMID: 40384986
- PMCID: PMC12079024
- DOI: 10.1002/mco2.70193
Minimal Residual Disease Detection: Implications for Clinical Diagnosis and Cancer Patient Treatment
Abstract
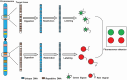
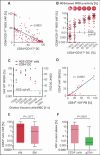
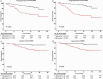
Minimal residual disease (MRD) serves as a pivotal biomarker for the clinical diagnosis and subsequent treatment of cancer patients. In hematological malignancies, MRD pose an increasingly serious threat to the health of Chinese people. Accurate MRD detection is essential for assessing relapse risk and optimizing therapeutic strategies, yet current methods such as flow cytometry, polymerase chain reaction (PCR), and next-generation sequencing (NGS) each have distinct limitations, and significant gaps remain in achieving optimal sensitivity and specificity of these technologies. This review provides a comprehensive analysis of MRD detection methods, high-lighting their clinical implications, including their roles in treatment decision-making, risk stratification, and patient outcomes. It discusses the strengths and weaknesses of existing techniques and explores emerging technologies that promise enhanced diagnostic precision. Key advancements such as integrating NGS with other methodologies and novel approaches like liquid biopsy and PCR are examined. The review underscores the academic and practical value of early and accurate MRD detection, emphasizing its impact on improving patient management and treatment outcomes. By addressing the limitations of current technologies and exploring future directions, this review aims to advance the field and support personalized medicine approaches to cancer treatment.
Keywords: MRD; clinical implication; detection methods; next‐generation sequencing.
© 2025 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Yang F., Anekpuritanang T., and Press R. D., “Clinical Utility of Next‐Generation Sequencing in Acute Myeloid Leukemia,” Molecular Diagnosis & Therapy 24, no. 1 (2020): 1–13. - PubMed
-
- Li W.. Measurable Residual Disease Testing in Acute Leukemia: Technology and Clinical Significance. In: Li W, ed. “Leukemia” (Brisbane (AU): Exon Publications, 2022). October 16,. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
