GLS1 promotes lipid metabolism in hepatocellular carcinoma by regulating the PI3K/AKT/mTORC1 signaling pathway through SREBP-1
- PMID: 40385002
- PMCID: PMC12082496
- DOI: 10.62347/ZTGP5030
GLS1 promotes lipid metabolism in hepatocellular carcinoma by regulating the PI3K/AKT/mTORC1 signaling pathway through SREBP-1
Abstract
Objectives: Cancer cells exhibit altered metabolic profiles. Glutaminase 1 (GLS1), a key enzyme in cancer cells, promoting glutamine catabolism to glutamate and ammonia, is strongly associated with various human malignancies.
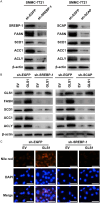
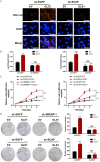
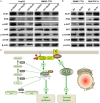
Methods: GLS1 promotes lipid accumulation and cell proliferation by upregulating the expression of sterol regulatory element-binding protein 1 (SREBP-1) and SREBP cleavage-activating protein (SCAP). Mechanistically, GLS1 promotes lipid metabolism in HCC cells through the activation of the PI3K/AKT/mTORC pathway.
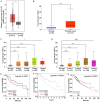
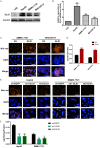
Results: GLS1's role in lipid metabolism in hepatocellular carcinoma (HCC) remains unexplored. Our findings indicate that GLS1 is not only significantly overexpressed in HCC but also negatively correlates with clinical prognosis. Further investigation revealed that GLS1 drives lipid accumulation and de novo fatty acid synthesis in HCC.
Conclusions: Our study suggests that GLS1 mediates SREBP-1 to drive lipid metabolism in HCC via the phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin complex 1 (PI3K/AKT/mTORC1) signaling pathway, thus we present GLS1 as a potential biomarker and therapeutic target for HCC.
Keywords: GLS1; Hepatocellular carcinoma; PI3K/AKT/mTORC1 signaling pathway; SREBP-1; lipid metabolism.
AJTR Copyright © 2025.
Conflict of interest statement
None.
Figures

Similar articles
-
GLS1 promotes proliferation in hepatocellular carcinoma cells via AKT/GSK3β/CyclinD1 pathway.Exp Cell Res. 2019 Aug 1;381(1):1-9. doi: 10.1016/j.yexcr.2019.04.005. Epub 2019 May 3. Exp Cell Res. 2019. PMID: 31054856
-
Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma.Acta Pharm Sin B. 2022 Feb;12(2):558-580. doi: 10.1016/j.apsb.2021.09.019. Epub 2021 Sep 25. Acta Pharm Sin B. 2022. PMID: 35256934 Free PMC article. Review.
-
Positive feedback loop between PI3K-Akt-mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen.Cancer Res. 2011 Jul 15;71(14):4989-97. doi: 10.1158/0008-5472.CAN-10-4108. Epub 2011 Jun 1. Cancer Res. 2011. PMID: 21632551
-
Pleiotrophin, a target of miR-384, promotes proliferation, metastasis and lipogenesis in HBV-related hepatocellular carcinoma.J Cell Mol Med. 2017 Nov;21(11):3023-3043. doi: 10.1111/jcmm.13213. Epub 2017 May 30. J Cell Mol Med. 2017. PMID: 28557334 Free PMC article.
-
Connecting mTORC1 signaling to SREBP-1 activation.Curr Opin Lipidol. 2012 Jun;23(3):226-234. doi: 10.1097/MOL.0b013e328352dd03. Curr Opin Lipidol. 2012. PMID: 22449814 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
