Integrating genome and transcriptome-wide data to explore the expression dynamics of TCP genes in Pisum sativum under salt stress
- PMID: 40385237
- PMCID: PMC12083428
- DOI: 10.3389/fpls.2025.1580890
Integrating genome and transcriptome-wide data to explore the expression dynamics of TCP genes in Pisum sativum under salt stress
Abstract
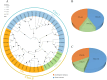
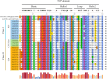
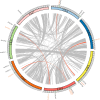
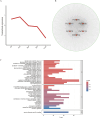
Salt stress severely restricts plant growth and productivity. TCP genes, which are plant-specific transcription factors, play a crucial role in the stress response. However, their functions in pea (Pisum sativum) remain poorly understood. Here, we identified 21 PsTCP genes in pea, classified into Class I (PCF) and Class II (CYC/TB1 and CIN) through phylogenetic analysis. While physicochemical properties varied significantly within the PsTCP family, gene structures and conserved motifs were highly conserved among subfamilies. Comparative homology analysis revealed closer relationships between pea TCP genes and dicots (Arabidopsis) than monocots (rice). Cis-regulatory element analysis suggested roles in growth, hormone response, and stress adaptation. Under salt stress, PsTCP genes exhibited divergent expression patterns, with PsTCP17 showing significant upregulation under extreme stress. Weighted gene co-expression network (WGCNA) and gene ontology (GO) enrichment analyses identified PsTCP20 as a hub gene regulating photosynthesis and metabolic processes. Tissue-specific expression across 11 pea tissues further highlighted their functional diversity. This study provides insights into the molecular mechanisms of salt stress responses in pea and offers genetic resources for breeding salt-tolerant varieties.
Keywords: Pisum sativum; TCP gene family; WGCNA; salt stress; tissue-specific expression.
Copyright © 2025 Fangyuan, Yong, Huang, Zhiyue and Wen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Amarakoon D., Thavarajah D., Mcphee K., Thavarajah P. (2012). Iron-, zinc-, and magnesium-rich field peas (Pisum sativum L.) with naturally low phytic acid: A potential food-based solution to global micronutrient malnutrition. J. Food Composition Anal. 27, 8–13. doi: 10.1016/j.jfca.2012.05.007 - DOI
-
- Ben Amor N., Jiménez A., Boudabbous M., Sevilla F., Abdelly C. (2020). Chloroplast implication in the tolerance to salinity of the halophyte Cakile maritima . Russian J. Plant Physiol. 67, 507–514. doi: 10.1134/S1021443720030048 - DOI
LinkOut - more resources
Full Text Sources

