Development of peripheral biomarker-based prognostic nomograms for short-term and long-term survival in immune checkpoint inhibitor-associated myocarditis
- PMID: 40385270
- PMCID: PMC12082187
- DOI: 10.21037/cdt-24-556
Development of peripheral biomarker-based prognostic nomograms for short-term and long-term survival in immune checkpoint inhibitor-associated myocarditis
Abstract
Background: Immune checkpoint inhibitor-associated myocarditis (ICI myocarditis) is a rare but highly fatal immune-related adverse reaction. This study aimed to develop nomogram prognostic models for both short-term and long-term survival outcomes in patients with ICI myocarditis based on key biomarkers in peripheral blood.
Methods: In this single-center retrospective study, we included 90 patients with ICI myocarditis at the Fourth Hospital of Hebei Medical University. Critical peripheral biomarkers associated with 40-day and 1-year overall survival (OS) were identified. Two prognostic models were developed and evaluation of the models were performed with receiver operating characteristic (ROC) curves, C-index, calibration curves, and decision curve analysis (DCA).
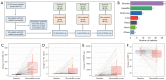
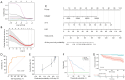
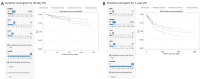
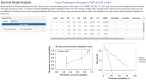
Results: A total of 24 patients (26.7%) succumbed within 40 days, while 40 patients (44.4%) died within one year. Cardiac troponin-I (cTnI), N-terminal pro-brain natriuretic peptide (NTBNP) and lactic dehydrogenase-to-albumin ratio (LAR) were identified as critical prognostic factors for 40-day OS in patients with ICI myocarditis and utilized to develop a nomogram model. The model demonstrates an area under the curve (AUC) of 0.867 [95% confidence interval (CI): 0.774-0.960] and a C-index of 0.824. Another predictive model for the 1-year OS was developed based on cTnI, NTBNP, LAR and systemic inflammatory response index (SIRI) with an AUC of 0.765 (95% CI: 0.664-0.866) and a C index of 0.742. The calibration curve demonstrates that both models exhibit strong consistency. The results of the DCA further indicate that both nomograms possess substantial clinical utility.
Conclusions: These two prediction models will enable clinicians to more effectively utilize readily available peripheral blood biomarkers for the convenient and efficient identification of high-risk patients with poor prognoses, thereby facilitating early intervention.
Keywords: Immune checkpoint inhibitor (ICI); biomarkers; myocarditis; nomogram; prognosis.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-24-556/coif). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Nomograms based on ratio indexes to predict severity and prognosis in immune checkpoint inhibitors-related myocarditis: a retrospective analysis.J Cancer Res Clin Oncol. 2024 May 27;150(5):277. doi: 10.1007/s00432-024-05801-7. J Cancer Res Clin Oncol. 2024. PMID: 38801421 Free PMC article.
-
Cardiovascular adverse events associated with immune checkpoint inhibitors: A retrospective multicenter cohort study.Cancer Med. 2024 May;13(10):e7233. doi: 10.1002/cam4.7233. Cancer Med. 2024. PMID: 38752474 Free PMC article.
-
Peripheral biomarkers to assess risk, severity, and prognosis of immune checkpoint inhibitor-associated myocarditis: a retrospective clinical study.Front Cardiovasc Med. 2024 Oct 24;11:1465743. doi: 10.3389/fcvm.2024.1465743. eCollection 2024. Front Cardiovasc Med. 2024. PMID: 39512372 Free PMC article.
-
Predictive value of inflammation and nutritional index in immunotherapy for stage IV non-small cell lung cancer and model construction.Sci Rep. 2024 Jul 30;14(1):17511. doi: 10.1038/s41598-024-66813-4. Sci Rep. 2024. PMID: 39080372 Free PMC article.
-
Efficacy of high-dose steroids versus low-dose steroids in the treatment of immune checkpoint inhibitor-associated myocarditis: a case series and systematic review.Front Immunol. 2025 Feb 12;16:1455347. doi: 10.3389/fimmu.2025.1455347. eCollection 2025. Front Immunol. 2025. PMID: 40013153 Free PMC article.
References
-
- Wei SC, Duffy CR, Allison JP. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov 2018;8:1069-86. 10.1158/2159-8290.CD-18-0367 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
