This is a preprint.
Integrative multi-omics QTL colocalization maps regulatory architecture in aging human brain
- PMID: 40385406
- PMCID: PMC12083576
- DOI: 10.1101/2025.04.17.25326042
Integrative multi-omics QTL colocalization maps regulatory architecture in aging human brain
Abstract
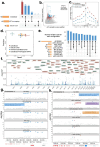
Multi-trait QTL (xQTL) colocalization has shown great promises in identifying causal variants with shared genetic etiology across multiple molecular modalities, contexts, and complex diseases. However, the lack of scalable and efficient methods to integrate large-scale multi-omics data limits deeper insights into xQTL regulation. Here, we propose ColocBoost, a multi-task learning colocalization method that can scale to hundreds of traits, while accounting for multiple causal variants within a genomic region of interest. ColocBoost employs a specialized gradient boosting framework that can adaptively couple colocalized traits while performing causal variant selection, thereby enhancing the detection of weaker shared signals compared to existing pairwise and multi-trait colocalization methods. We applied ColocBoost genome-wide to 17 gene-level single-nucleus and bulk xQTL data from the aging brain cortex of ROSMAP individuals (average ), encompassing 6 cell types, 3 brain regions and 3 molecular modalities (expression, splicing, and protein abundance). Across molecular xQTLs, ColocBoost identified 16,503 distinct colocalization events, exhibiting 10.7(±0.74)-fold enrichment for heritability across 57 complex diseases/traits and showing strong concordance with element-gene pairs validated by CRISPR screening assays. When colocalized against Alzheimer's disease (AD) GWAS, ColocBoost identified up to 2.5-fold more distinct colocalized loci, explaining twice the AD disease heritability compared to fine-mapping without xQTL integration. This improvement is largely attributable to ColocBoost's enhanced sensitivity in detecting gene-distal colocalizations, as supported by strong concordance with known enhancer-gene links, highlighting its ability to identify biologically plausible AD susceptibility loci with underlying regulatory mechanisms. Notably, several genes including BLNK and CTSH showed sub-threshold associations in GWAS, but were identified through multi-omics colocalizations which provide new functional support for their involvement in AD pathogenesis.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures





Similar articles
-
Joint analysis of GWAS and multi-omics QTL summary statistics reveals a large fraction of GWAS signals shared with molecular phenotypes.Cell Genom. 2023 Jun 19;3(8):100344. doi: 10.1016/j.xgen.2023.100344. eCollection 2023 Aug 9. Cell Genom. 2023. PMID: 37601976 Free PMC article.
-
COLOCdb: a comprehensive resource for multi-model colocalization of complex traits.Nucleic Acids Res. 2024 Jan 5;52(D1):D871-D881. doi: 10.1093/nar/gkad939. Nucleic Acids Res. 2024. PMID: 37941154 Free PMC article.
-
Identifying cross-tissue molecular targets of lung function by multi-omics integration analysis from DNA methylation and gene expression of diverse human tissues.BMC Genomics. 2025 Mar 24;26(1):289. doi: 10.1186/s12864-025-11476-2. BMC Genomics. 2025. PMID: 40128644 Free PMC article.
-
From GWAS to Gene: Transcriptome-Wide Association Studies and Other Methods to Functionally Understand GWAS Discoveries.Front Genet. 2021 Sep 30;12:713230. doi: 10.3389/fgene.2021.713230. eCollection 2021. Front Genet. 2021. PMID: 34659337 Free PMC article. Review.
-
From genetic associations to genes: methods, applications, and challenges.Trends Genet. 2024 Aug;40(8):642-667. doi: 10.1016/j.tig.2024.04.008. Epub 2024 May 11. Trends Genet. 2024. PMID: 38734482 Review.
References
-
- Bryois J. et al. Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nature neuroscience 25, 1104–1112 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
