This is a preprint.
Androgens mediate sexual dimorphism in Pilarowski-Bjornsson Syndrome
- PMID: 40385454
- PMCID: PMC12083630
- DOI: 10.1101/2025.05.06.25326635
Androgens mediate sexual dimorphism in Pilarowski-Bjornsson Syndrome
Abstract
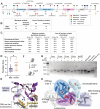
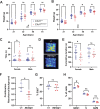
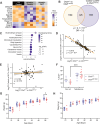
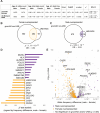
Sex-specific penetrance in autosomal dominant Mendelian conditions is largely understudied. The neurodevelopmental disorder Pilarowski-Bjornsson syndrome (PILBOS) was initially described in females. Here, we describe the clinical and genetic characteristics of the largest PILBOS cohort to date, showing that both sexes can exhibit PILBOS features, although males are overrepresented. A mouse model carrying a human-derived Chd1 missense variant (Chd1 R616Q/+) displays female-restricted phenotypes, including growth deficiency, anxiety and hypotonia. Orchiectomy unmasks a growth deficiency phenotype in male Chd1 R616Q/+ mice, while testosterone rescues the phenotype in females, implicating androgens in phenotype modulation. In the gnomAD and UK Biobank databases, rare missense variants in CHD1 are overrepresented in males, supporting a male protective effect. We identify 33 additional highly constrained autosomal genes with missense variant overrepresentation in males. Our results support androgen-regulated sexual dimorphism in PILBOS and open novel avenues to understand the mechanistic basis of sexual dimorphism in other autosomal Mendelian disorders.
Keywords: CHD1; Mendelian disease; Neurodevelopmental disorder; Sex differences.
Conflict of interest statement
Competing interests Dr. Bjornsson is Founder of KALDUR therapeutics and a consultant for Mahzi therapeutics. S.B. is a part of a working group called Alzheimer diagnostics that has received a grant from the Icelandic Technology Development Fund. I.M.W. is an employee of and may own stock in GeneDx. E.E.E. is a scientific advisory board (SAB) member of Variant Bio, Inc. K.S. and A.H. are employees of deCODE genetics, a subsidiary of Amgen. The Department of Medical and Molecular Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories.
Figures

References
-
- Sunwoo Y., Seo S. H., Kim H.-J., Park M. S. & Cho A. Variant of CHD1 gene resulting in a Korean case of Pilarowski-Bjornsson syndrome. J Genet Med 19, 111–114 (2022). 10.5734/JGM.2022.19.2.111 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
