Identification and genomic characterisation of known and novel highly divergent sapoviruses in frugivorous and insectivorous bats in Nigeria
- PMID: 40385501
- PMCID: PMC12080456
- DOI: 10.1080/29986990.2025.2503155
Identification and genomic characterisation of known and novel highly divergent sapoviruses in frugivorous and insectivorous bats in Nigeria
Abstract
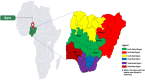
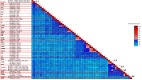
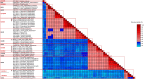
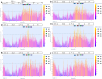
Sapovirus (SaV) infections have been linked with moderate-to-severe acute gastroenteritis (AGE) in animals and humans and represent a significant risk to public health. SaVs from animals including pigs, chimpanzees, and rodents have been reported to be closely related with human SaVs, indicating the possibility of cross-species transmission. Divergent SaVs have been reported in various bat species across various continents including Asia, Europe, Oceania and Africa. However, little is known about the evolutionary history of SaVs across various bat species and their zoonotic potential. In this report, we describe the findings of a surveillance study across various bat species in Nigeria. Samples were pooled and subjected to metagenomics sequencing and analyses. Nine of 57 sample pools (containing 223 rectal swabs from five bat species) had SaV reads from which we assembled a total of four complete and three near-complete (having complete coding sequences) genomes. The bat SaV (BtSaV) strains from this study formed five distinct lineages of which four represented novel genogroups. BtSaV lineages clustered mainly according to bat families, which might suggest a likely virus-host-specific evolution. The BtSaV VP1 capsid protein structure prediction confirmed three main domains (S, P1, and P2) as reported for Human SaV (HuSaV). We found that the P2 subdomain of the VP1 protein contains a degree of homology to known immunoreactive epitopes suggesting these conserved regions may be valuable for diagnostics or medical countermeasure development. This study expands our understanding of reservoir hosts, provides information on the genetic diversity and continuous evolution of SaVs in bats.
Keywords: Nigeria; Sapovirus; VP1 protein; bats; metagenomic sequencing; new genogroups; protein modelling.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Ten Previously Unassigned Human Cosavirus Genotypes Detected in Feces of Children with Non-Polio Acute Flaccid Paralysis in Nigeria in 2020.Viruses. 2025 Jun 12;17(6):844. doi: 10.3390/v17060844. Viruses. 2025. PMID: 40573435 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
