Impact of different genomic relationship matrix construction methods on the accuracy of genomic prediction in different species
- PMID: 40385981
- PMCID: PMC12082045
- DOI: 10.3389/fgene.2025.1576248
Impact of different genomic relationship matrix construction methods on the accuracy of genomic prediction in different species
Abstract
Objective: Genomic best linear unbiased prediction (GBLUP) is a key method in genomic prediction, relying on the construction of a genomic relationship matrix (G-matrix). Although various methods for G-matrix construction have been proposed, the performance of these methods across different species has not been thoroughly compared.
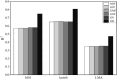
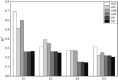
Methods: This study systematically evaluated the performance of six genomic relationship matrix (G-matrix) construction methods in improving the prediction accuracy of GBLUP models across four species: pigs, bulls, wheat, and mice. The methodological framework included: (1) an initial unscaled matrix; (2) five scaled methods utilizing allele frequency centralization. The scaled methods comprised: (a) three variance-weighted approaches using allele frequencies fixed at 0.5 (G05), observed frequencies (GOF), or average minor allele frequencies (GMF); (b) two centralized methods with weighting by either the trace of the numerator matrix (GN) or reciprocals of each locus's expected variance (GD).
Results: The GD matrix demonstrated significant prediction accuracy improvements for pig traits. Conversely, most scaled G-matrices showed minimal effects on mice, wheat, and bull, even with underperforming unscaled baselines in prediction accuracy compared to the original unscaled matrix. The learning curve for bull data showed the choice of G-matrix had minimal impact on prediction accuracy when the reference population size and genetic marker density reached a certain threshold.
Discussion: The study concluded that the optimal G-matrix construction method varies across species, with population structure being a key factor. These findings highlight the importance of species-specific optimization in genomic prediction and suggest that the influence of G-matrix construction diminishes in large-scale, high-density genomic datasets.
Keywords: accuracy of prediction; different species; genomic relationship matrix; marker density; size of reference population.
Copyright © 2025 Wang, Wei, Liu, Zhang, Wang, Pan and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Different genomic relationship matrices for single-step analysis using phenotypic, pedigree and genomic information.Genet Sel Evol. 2011 Jan 5;43(1):1. doi: 10.1186/1297-9686-43-1. Genet Sel Evol. 2011. PMID: 21208445 Free PMC article.
-
Comparison of genomic predictions using genomic relationship matrices built with different weighting factors to account for locus-specific variances.J Dairy Sci. 2014 Oct;97(10):6547-59. doi: 10.3168/jds.2014-8210. Epub 2014 Aug 14. J Dairy Sci. 2014. PMID: 25129495
-
Improving Genomic Predictions in Multi-Breed Cattle Populations: A Comparative Analysis of BayesR and GBLUP Models.Genes (Basel). 2024 Feb 18;15(2):253. doi: 10.3390/genes15020253. Genes (Basel). 2024. PMID: 38397242 Free PMC article.
-
Efficient weighting methods for genomic best linear-unbiased prediction (BLUP) adapted to the genetic architectures of quantitative traits.Heredity (Edinb). 2021 Feb;126(2):320-334. doi: 10.1038/s41437-020-00372-y. Epub 2020 Sep 26. Heredity (Edinb). 2021. PMID: 32980863 Free PMC article.
-
The effect of high-density genotypic data and different methods on joint genomic prediction: A case study in large white pigs.Anim Genet. 2023 Feb;54(1):45-54. doi: 10.1111/age.13275. Epub 2022 Nov 22. Anim Genet. 2023. PMID: 36414135
References
LinkOut - more resources
Full Text Sources

