Chromosome-level draft genome assembly of Hypomesus nipponensis reveals transposable element expansion reshaping the genome structure
- PMID: 40385984
- PMCID: PMC12083275
- DOI: 10.3389/fgene.2025.1502681
Chromosome-level draft genome assembly of Hypomesus nipponensis reveals transposable element expansion reshaping the genome structure
Abstract
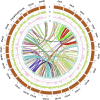
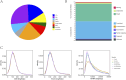
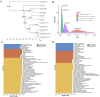
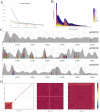
Hypomesus nipponensis a commercially valuable fish within the Osmeriformes order, is naturally found in northeastern Asia and has been extensively introduced for commercial purposes across eastern Asia. To investigate the taxonomic status and evolutionary history of Hypomesus nipponensis within the Osmeridae family, we first performed a de novo genome assembly using PacBio HiFi reads and CLR (Continuous Long Read) reads. Subsequently, we leveraged synteny information from closely related species to further refine the assembly and construct a chromosome-level genome. The final assembly spans 507.8 Mb, with a scaffold N50 of 20 Mb, achieving chromosome-level contiguity. It comprises 164 Mb of repetitive sequences and encodes 27,876 protein-coding genes. Compared to previous assembly, the H. nipponensis genome is notably more contiguous and complete. Notably, it contains an unusually high proportion of tandem repeats, which likely contributed to the assembly challenges encountered in earlier efforts. We also observed the transposons of H. nipponensis have expanded significantly in recent times, and paralogous gene families have expanded during the same period. Our analysis estimates that H. nipponensis, Osmerus eperlanus, and Hypomesus transpacificus diverged from a common ancestor approximately 24.1 million years ago, with significant chromosomal segment recombination events occurring during their divergence. Additionally, we compared the genomes of O. eperlanus and Hypomesus and found that most of the genes in the Presence/Absence Variants (PAVs) of O. eperlanus were associated with immune response. Our efforts significantly enhance the genome's integrity and continuity for this ecologically and commercially important fish, providing a chromosome-level genome draft that supports fundamental biological research while offering insights into the evolutionary relationships and genomic diversity within the Osmeriformes order. This advancement has profound implications for understanding the evolutionary history and adaptive strategies of H. nipponensis.
Keywords: Hypomesus nipponensis; evolutionary; gene family; genome; repeat sequence.
Copyright © 2025 Zhu, Kuang, Li and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Complete mitochondrial genome of the surf smelt Hypomesus japonicus (Osmeriformes, Osmeridae).Mitochondrial DNA B Resour. 2018 Sep 10;3(2):1071-1072. doi: 10.1080/23802359.2018.1511852. Mitochondrial DNA B Resour. 2018. PMID: 33474419 Free PMC article.
-
Draft genome of the Korean smelt Hypomesus nipponensis and its transcriptomic responses to heat stress in the liver and muscle.G3 (Bethesda). 2021 Sep 6;11(9):jkab147. doi: 10.1093/g3journal/jkab147. G3 (Bethesda). 2021. PMID: 33944944 Free PMC article.
-
Genome sequence of the barred knifejaw Oplegnathus fasciatus (Temminck & Schlegel, 1844): the first chromosome-level draft genome in the family Oplegnathidae.Gigascience. 2019 Mar 1;8(3):giz013. doi: 10.1093/gigascience/giz013. Gigascience. 2019. PMID: 30715332 Free PMC article.
-
Complete mitochondrial genome of the European smelt Osmerus eperlanus (Osmeriformes, Osmeridae).Mitochondrial DNA B Resour. 2018 Jul 10;3(2):744-745. doi: 10.1080/23802359.2018.1483768. Mitochondrial DNA B Resour. 2018. PMID: 33474308 Free PMC article.
-
Complete mitochondrial genome of the Arctic rainbow smelt Osmerus dentex (Osmeriformes, Osmeridae).Mitochondrial DNA B Resour. 2018 Aug 27;3(2):879-880. doi: 10.1080/23802359.2018.1501301. Mitochondrial DNA B Resour. 2018. PMID: 33474351 Free PMC article.
References
-
- Asami H. (2004). Early life ecology of Japanese smelt (Hypomesus nipponensis) in Lake Abashiri, a brackish water, eastern Hokkaido, Japan. Sci. Rep. Hokkaido Fish. Res. Inst. (67).
LinkOut - more resources
Full Text Sources
Miscellaneous

